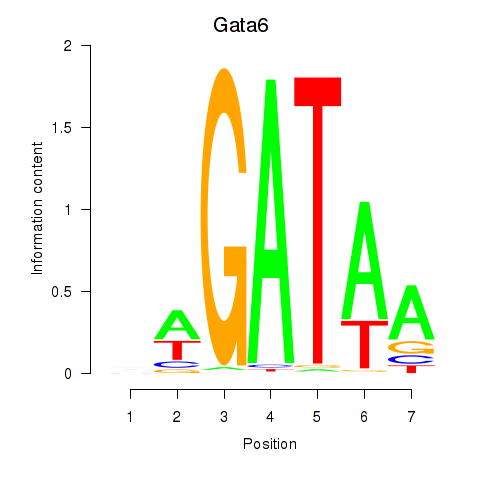
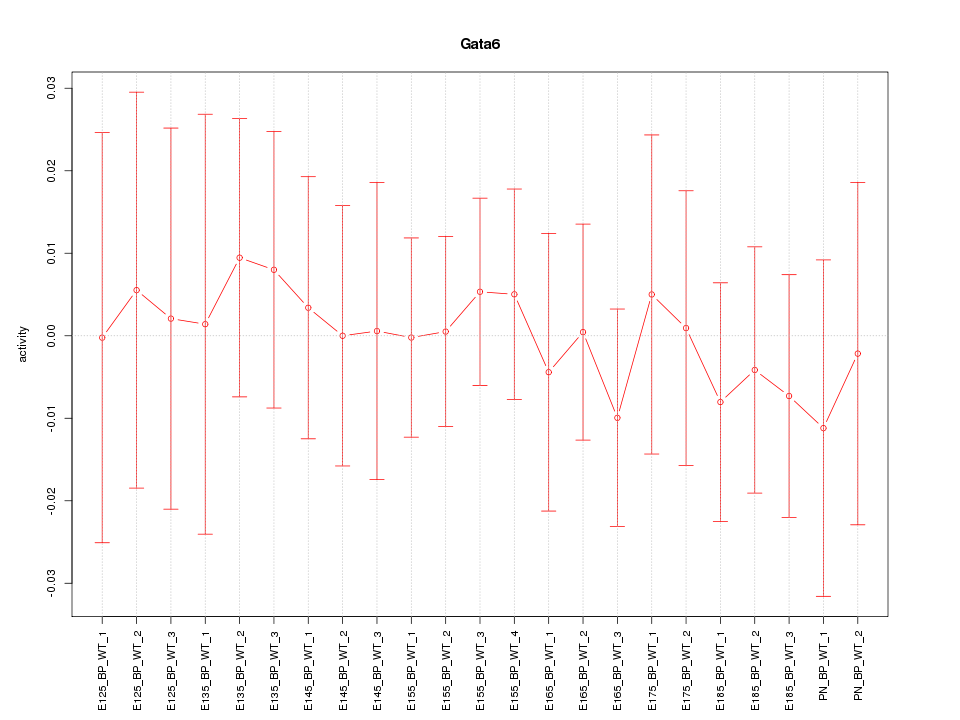
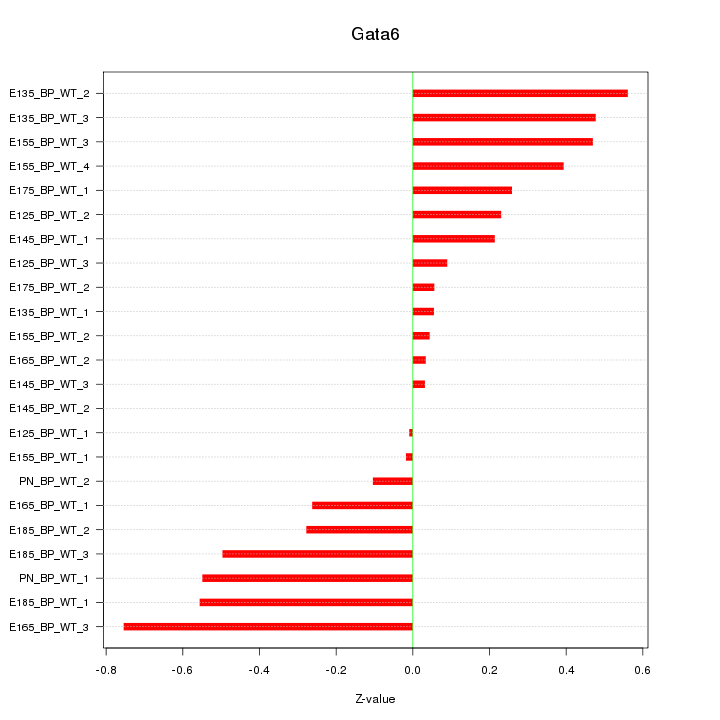
| 0.3 |
1.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
1.6 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
0.4 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.3 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 |
1.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.1 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 |
0.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.2 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.1 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.1 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.2 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.9 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.4 |
GO:0021692 |
cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.1 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |