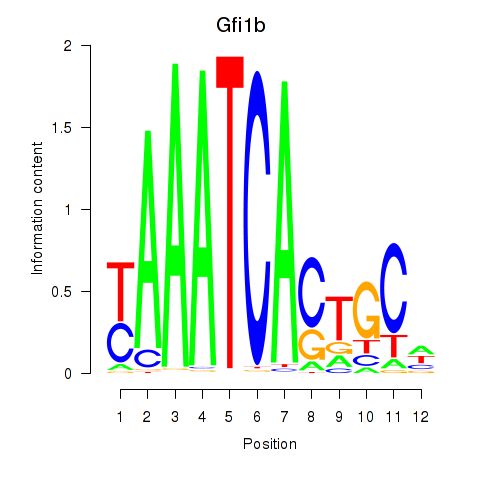
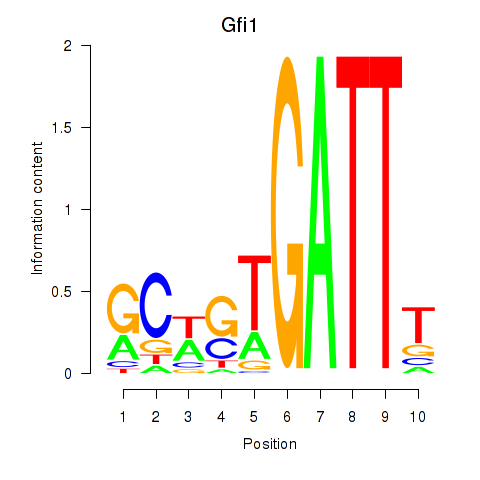
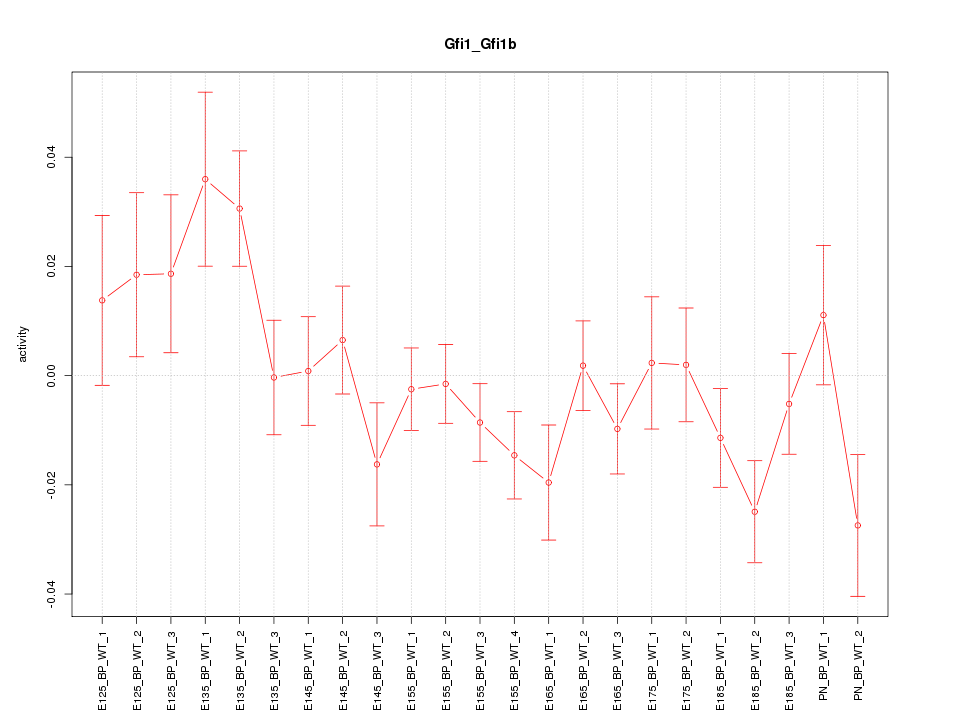
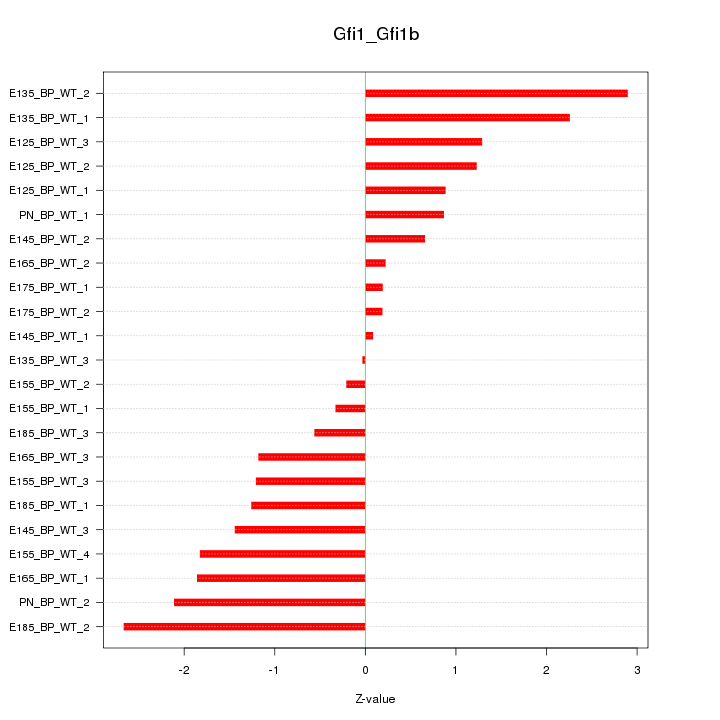
| 3.3 |
19.5 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.3 |
3.8 |
GO:0060067 |
cervix development(GO:0060067) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 1.0 |
2.9 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.7 |
2.1 |
GO:1990705 |
cholangiocyte proliferation(GO:1990705) |
| 0.7 |
2.0 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.5 |
2.1 |
GO:1900623 |
positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.5 |
1.5 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.5 |
1.5 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 |
0.5 |
GO:0071335 |
hair follicle cell proliferation(GO:0071335) |
| 0.5 |
2.3 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.5 |
9.2 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.4 |
1.3 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.4 |
0.8 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.4 |
2.7 |
GO:1900164 |
sequestering of extracellular ligand from receptor(GO:0035581) nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.4 |
1.1 |
GO:1902336 |
neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.4 |
1.1 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.4 |
1.1 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.3 |
0.9 |
GO:0046881 |
sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 |
0.9 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 |
1.5 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 |
1.5 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 |
9.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.3 |
0.8 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 |
6.6 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.3 |
0.8 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.3 |
1.6 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 |
1.5 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.2 |
0.7 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 |
0.2 |
GO:0060298 |
positive regulation of sarcomere organization(GO:0060298) |
| 0.2 |
0.7 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 |
0.9 |
GO:0070942 |
neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.2 |
1.7 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.2 |
0.8 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.2 |
1.6 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 |
0.7 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.2 |
0.9 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 |
1.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.2 |
0.5 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.2 |
1.8 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.2 |
0.5 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 |
2.5 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.2 |
0.6 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 |
2.4 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.2 |
1.0 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.2 |
0.5 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 |
0.8 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 |
1.8 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.9 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.4 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.3 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.4 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
1.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.7 |
GO:0060903 |
regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.4 |
GO:0032240 |
RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 |
0.4 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.6 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
2.0 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 |
0.7 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.6 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.1 |
0.1 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.1 |
0.2 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.1 |
0.4 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 |
0.8 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
0.9 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.4 |
GO:1904749 |
rRNA export from nucleus(GO:0006407) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 |
0.3 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
0.4 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
1.0 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.4 |
GO:0055009 |
extraocular skeletal muscle development(GO:0002074) atrial cardiac muscle tissue development(GO:0003228) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) atrial cardiac muscle tissue morphogenesis(GO:0055009) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 |
0.3 |
GO:2000768 |
positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 |
0.5 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 |
1.5 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
0.3 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.1 |
1.3 |
GO:0051382 |
protein localization to kinetochore(GO:0034501) kinetochore assembly(GO:0051382) |
| 0.1 |
0.2 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 |
0.7 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.2 |
GO:0045414 |
regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 |
1.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.1 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
1.3 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
2.3 |
GO:0000578 |
embryonic axis specification(GO:0000578) |
| 0.1 |
0.4 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
0.4 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 |
0.3 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.3 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
0.8 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
1.8 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 |
0.4 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.3 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.7 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.4 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
0.2 |
GO:0060160 |
musculoskeletal movement, spinal reflex action(GO:0050883) negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 |
1.5 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 |
1.5 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.1 |
GO:0097168 |
mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 |
0.2 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
1.4 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.1 |
2.3 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.0 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 |
0.1 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.1 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
1.2 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.6 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.5 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
1.8 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.2 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.3 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
1.0 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.7 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
1.8 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0060364 |
frontal suture morphogenesis(GO:0060364) |
| 0.0 |
0.2 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
1.4 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.3 |
GO:0016114 |
terpenoid biosynthetic process(GO:0016114) |
| 0.0 |
0.6 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.2 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.5 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
1.8 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.2 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.6 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 |
2.7 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.4 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
1.4 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.2 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 |
1.7 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.3 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.1 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.3 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.5 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.0 |
0.2 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.9 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.1 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.0 |
0.5 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.3 |
GO:0019731 |
antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.0 |
0.5 |
GO:0071353 |
response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 |
1.2 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.2 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.0 |
0.6 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.2 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.0 |
0.7 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.2 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
2.5 |
GO:0007612 |
learning(GO:0007612) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.2 |
GO:1903894 |
regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 |
0.7 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
1.1 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.3 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 |
0.3 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.1 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
1.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.0 |
GO:0015793 |
glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 |
0.1 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.3 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
1.0 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.0 |
0.3 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.8 |
GO:0031638 |
zymogen activation(GO:0031638) |
| 0.0 |
0.4 |
GO:0007091 |
metaphase/anaphase transition of mitotic cell cycle(GO:0007091) metaphase/anaphase transition of cell cycle(GO:0044784) |
| 0.0 |
0.3 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.3 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.1 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |