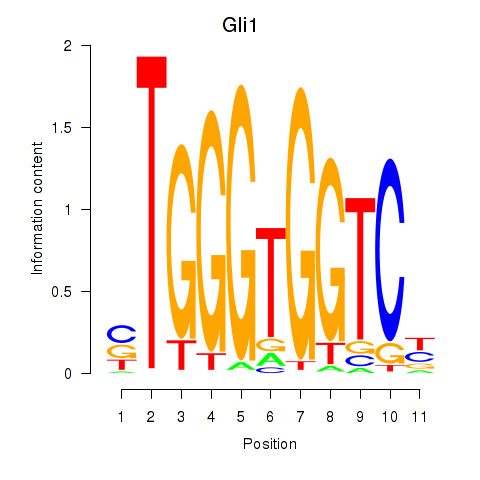
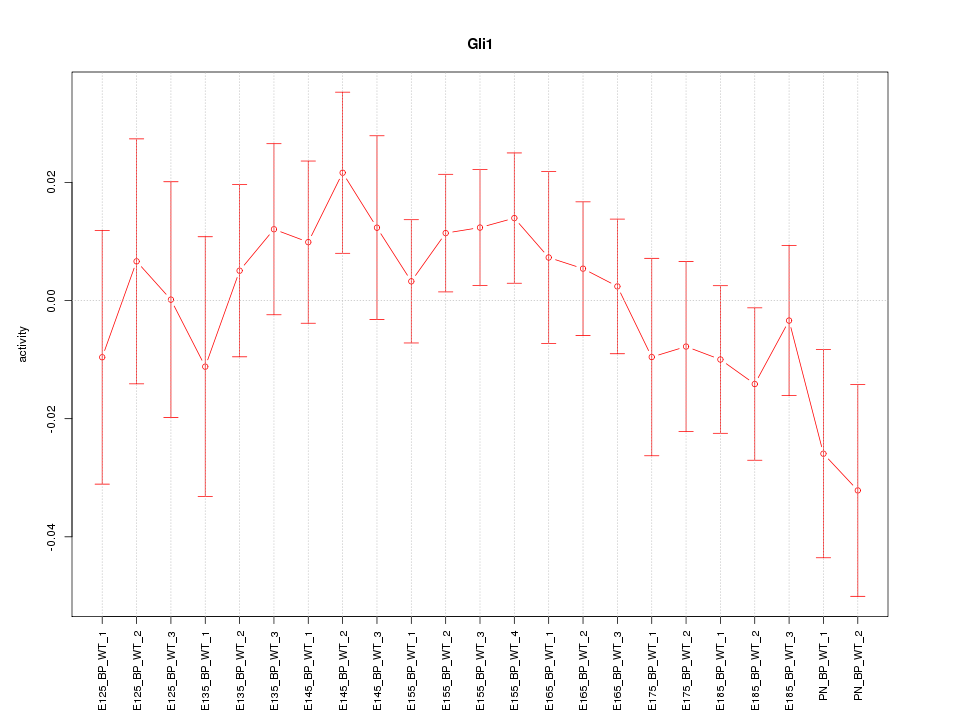
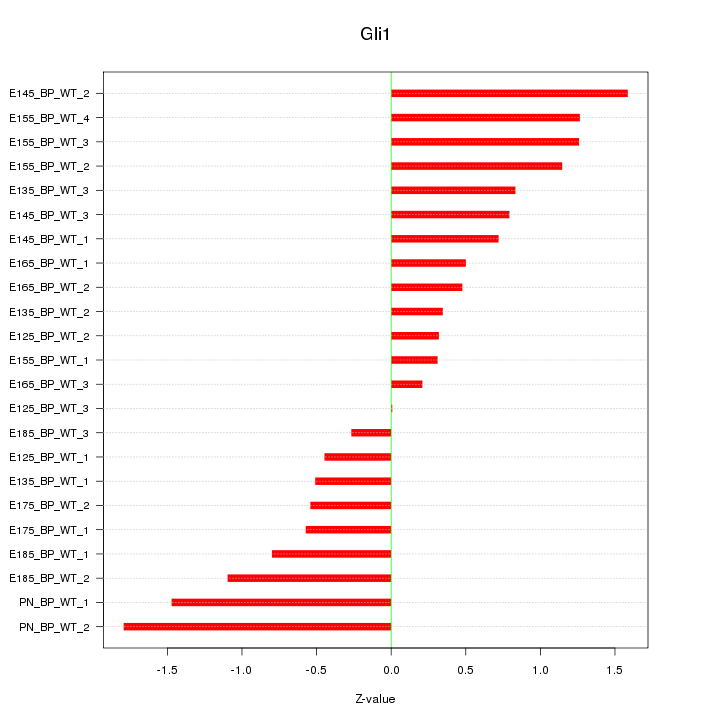
| 0.8 |
3.2 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.7 |
2.2 |
GO:0010751 |
negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.7 |
2.0 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.6 |
1.8 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
2.3 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.3 |
0.9 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.3 |
0.3 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.2 |
2.7 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.2 |
1.3 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 |
0.6 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 |
2.9 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 |
1.7 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 |
0.6 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.2 |
1.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.2 |
0.9 |
GO:1900451 |
rhythmic synaptic transmission(GO:0060024) terminal button organization(GO:0072553) positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of protein localization to synapse(GO:1902474) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 |
0.4 |
GO:0071336 |
lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 |
0.7 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
2.1 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.5 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
1.0 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.1 |
1.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.4 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 |
1.3 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
1.1 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.3 |
GO:0070671 |
response to interleukin-12(GO:0070671) |
| 0.1 |
2.6 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
1.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.5 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.2 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.2 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 |
1.6 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 |
0.3 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.1 |
2.5 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.1 |
0.2 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.1 |
0.2 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.3 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.7 |
GO:1901898 |
regulation of cell communication by electrical coupling(GO:0010649) negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.3 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.2 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 |
2.4 |
GO:1902017 |
regulation of cilium assembly(GO:1902017) |
| 0.0 |
0.7 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.5 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.3 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.2 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.5 |
GO:0030336 |
negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 |
0.2 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.4 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.1 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.0 |
0.3 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.4 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
0.8 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 |
0.1 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.6 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.4 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.6 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
2.5 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
1.0 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.3 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.7 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
1.6 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.1 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.1 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 0.0 |
0.5 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.0 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.0 |
0.4 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
1.3 |
GO:0002757 |
immune response-activating signal transduction(GO:0002757) |
| 0.0 |
0.7 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |