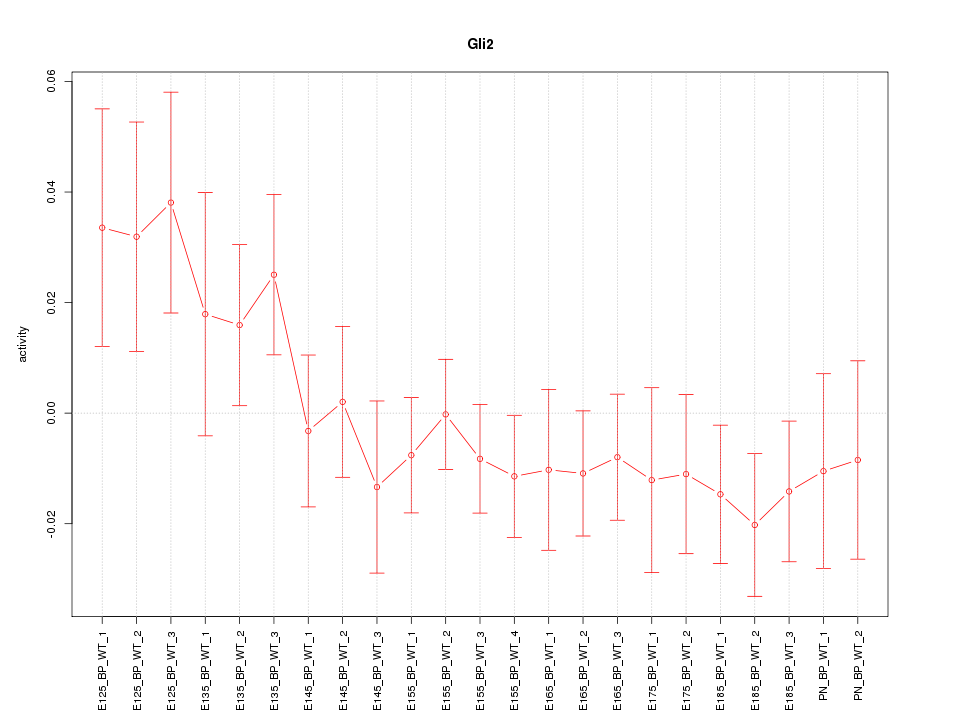
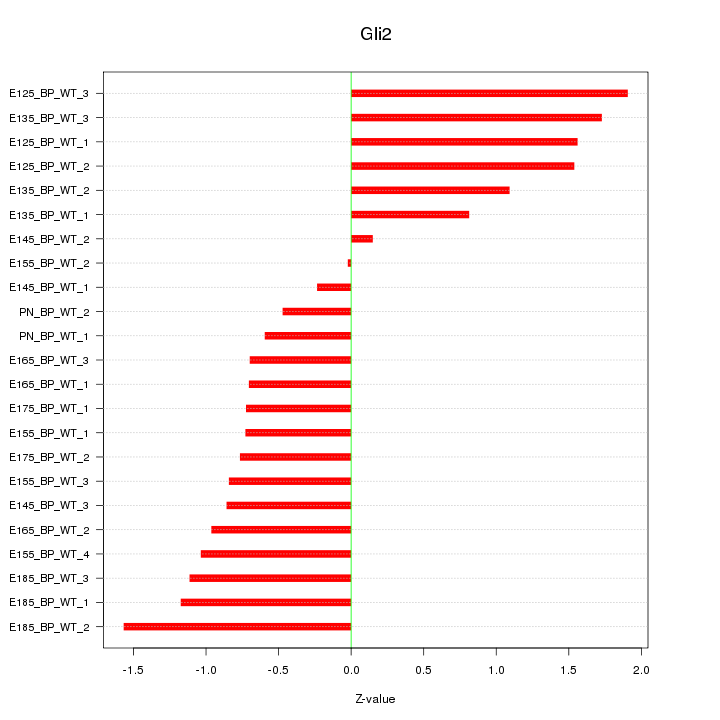
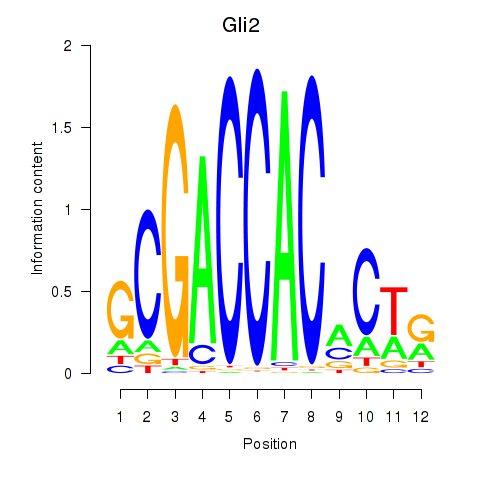
Motif ID: Gli2
Z-value: 1.044
Transcription factors associated with Gli2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gli2 | ENSMUSG00000048402.8 | Gli2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli2 | mm10_v2_chr1_-_119053339_119053370 | -0.09 | 6.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.8 | 7.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.4 | 8.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.1 | 6.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 1.9 | GO:0072070 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.4 | 1.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.4 | 2.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.4 | 1.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 1.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 3.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 | 0.8 | GO:0042713 | negative regulation of urine volume(GO:0035811) sperm ejaculation(GO:0042713) negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 0.8 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 1.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 1.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 4.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 1.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.4 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 0.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.2 | 0.7 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 0.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 2.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 4.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 2.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 2.1 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.2 | GO:1903960 | negative regulation of anion transmembrane transport(GO:1903960) |
| 0.0 | 0.2 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 5.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.8 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 2.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 1.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 2.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 5.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.7 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.2 | 3.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.1 | GO:0032281 | voltage-gated calcium channel complex(GO:0005891) AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 7.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.5 | 4.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.6 | 1.9 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.5 | 1.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.5 | 1.4 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.4 | 1.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 2.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 1.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 8.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 2.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 6.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 8.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 2.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 13.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |