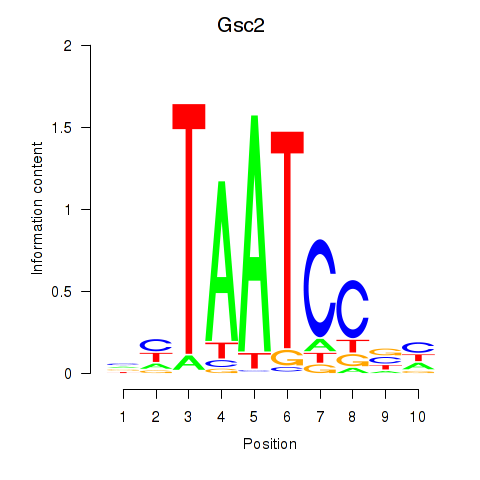
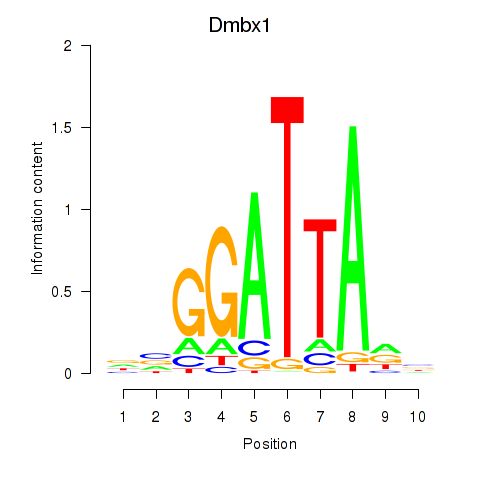
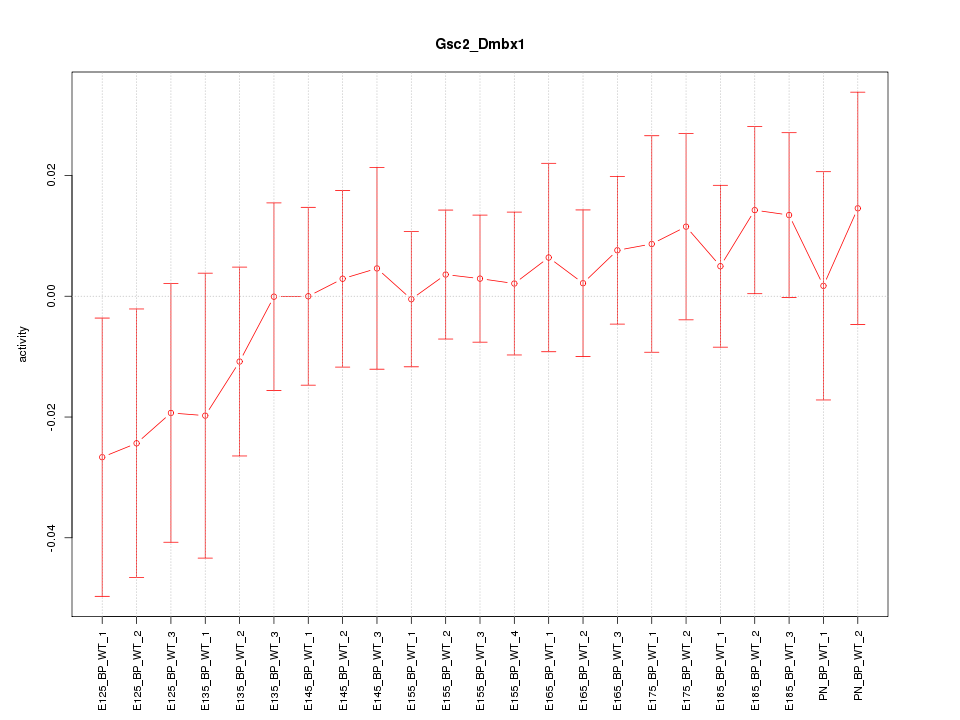
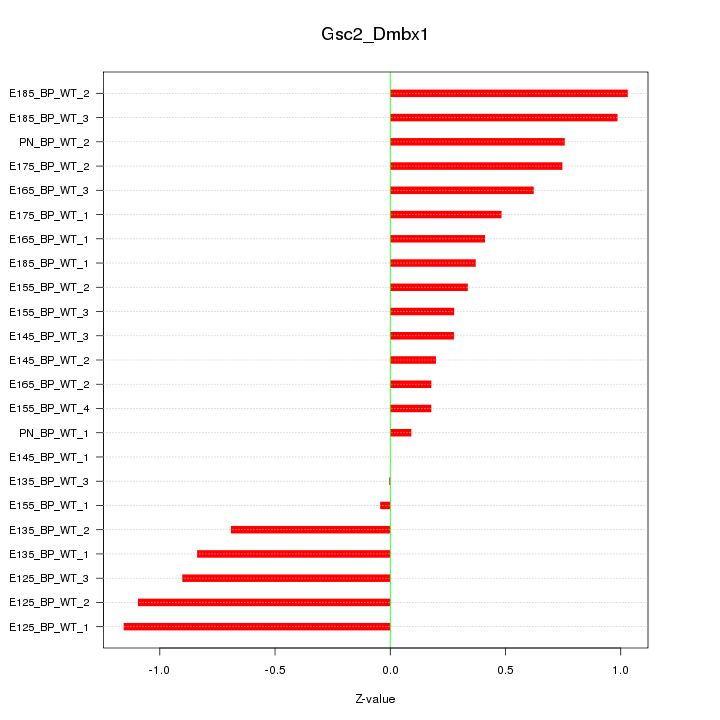
| 3.2 |
9.7 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 |
1.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.3 |
2.1 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.3 |
0.9 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 |
0.7 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 |
1.1 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 |
1.4 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.2 |
0.6 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
0.8 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
1.1 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
1.7 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.1 |
0.9 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.1 |
3.3 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 |
1.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
1.4 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 |
1.0 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
1.6 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.4 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
0.5 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
5.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
0.2 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 |
0.5 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:0009182 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.0 |
0.3 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.2 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.3 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.4 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.3 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
2.9 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 |
1.1 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
1.3 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.0 |
0.3 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.2 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.1 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.4 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |