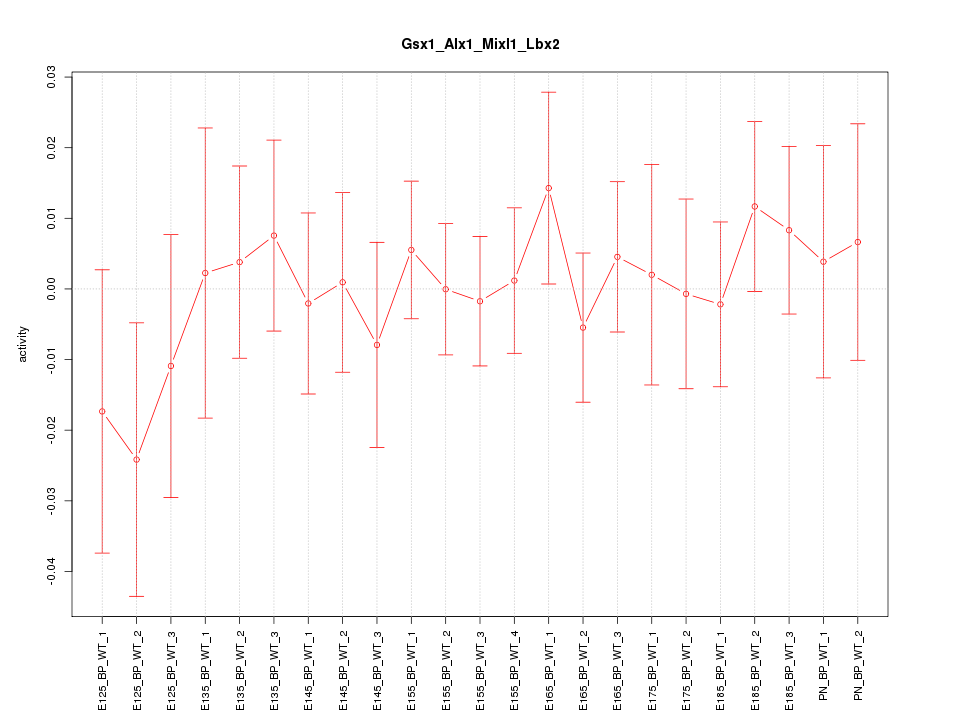
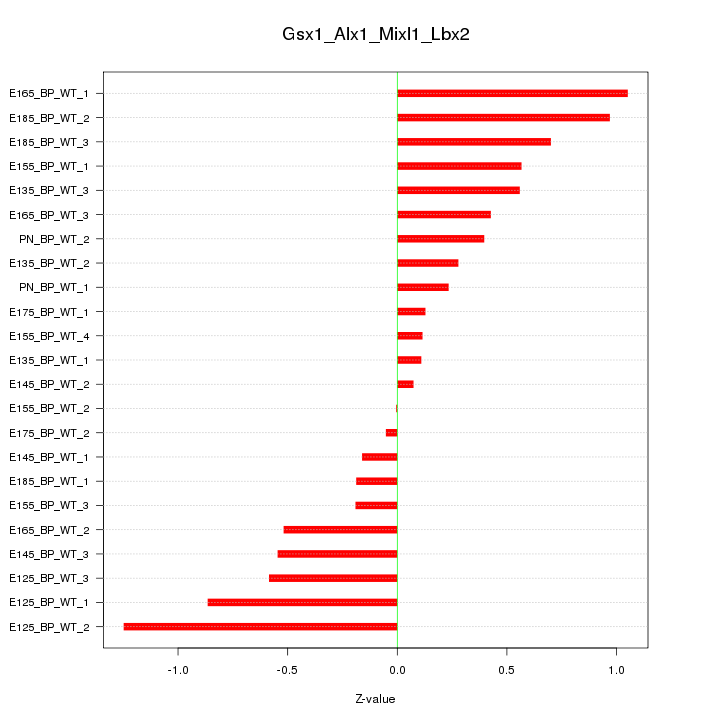
Motif ID: Gsx1_Alx1_Mixl1_Lbx2
Z-value: 0.552
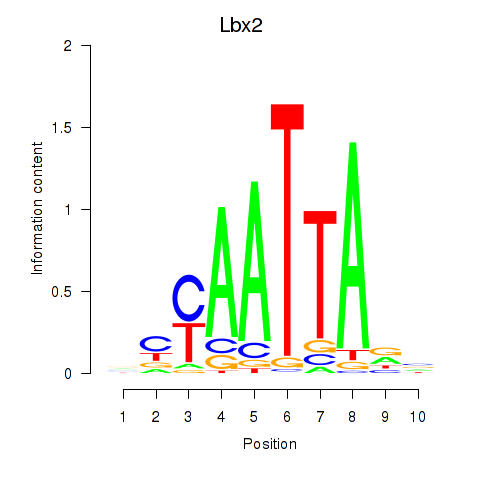
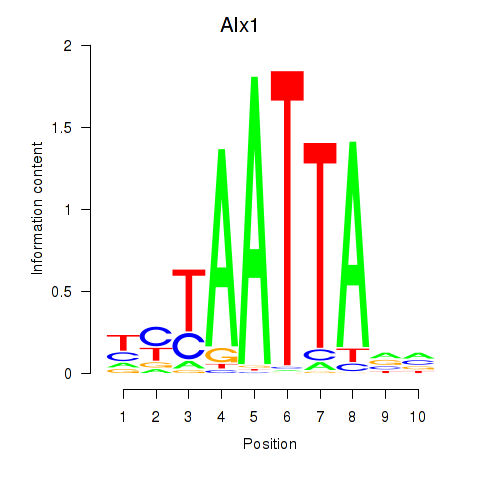
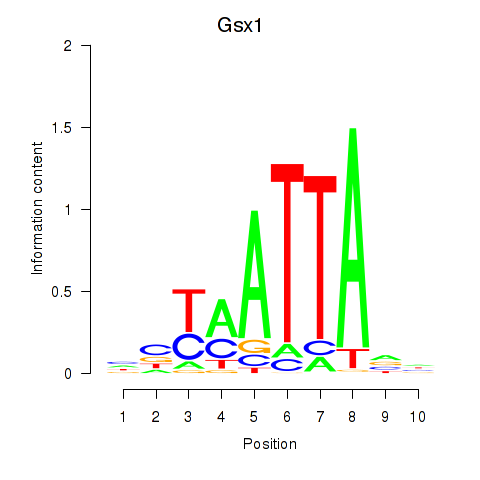
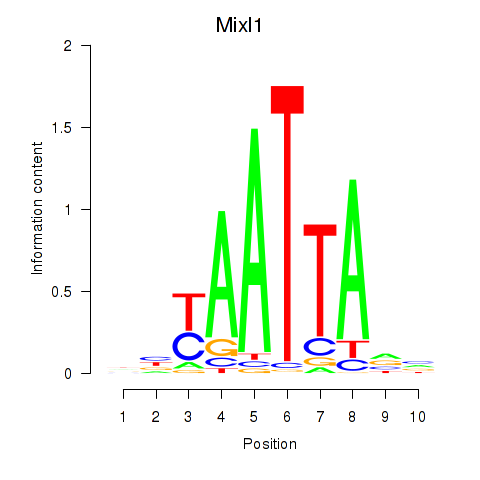
Transcription factors associated with Gsx1_Alx1_Mixl1_Lbx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Alx1 | ENSMUSG00000036602.7 | Alx1 |
| Gsx1 | ENSMUSG00000053129.5 | Gsx1 |
| Lbx2 | ENSMUSG00000034968.2 | Lbx2 |
| Mixl1 | ENSMUSG00000026497.7 | Mixl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx1 | mm10_v2_chr10_-_103028771_103028782 | 0.27 | 2.1e-01 | Click! |
| Gsx1 | mm10_v2_chr5_+_147188678_147188696 | 0.24 | 2.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.0 | 3.0 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.6 | 1.8 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 3.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 | 1.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 1.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.3 | 1.4 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 5.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.7 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.2 | 0.7 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 0.7 | GO:2001139 | negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.2 | 0.7 | GO:0071336 | lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 | 0.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.7 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 0.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 1.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 3.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 0.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.4 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.1 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 | 0.7 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 1.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 1.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.7 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 1.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:1903546 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.3 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 0.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 1.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 3.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 3.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.7 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 0.7 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 6.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 1.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 1.7 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 5.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |