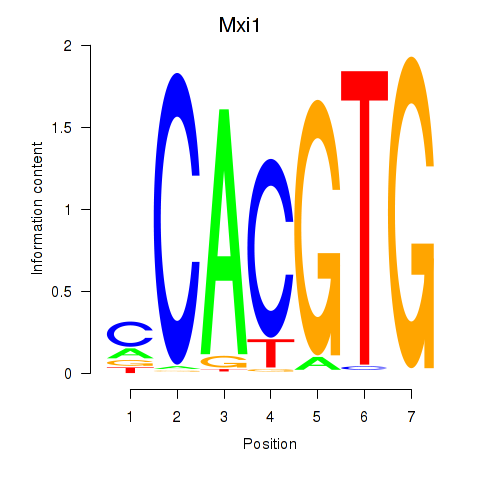
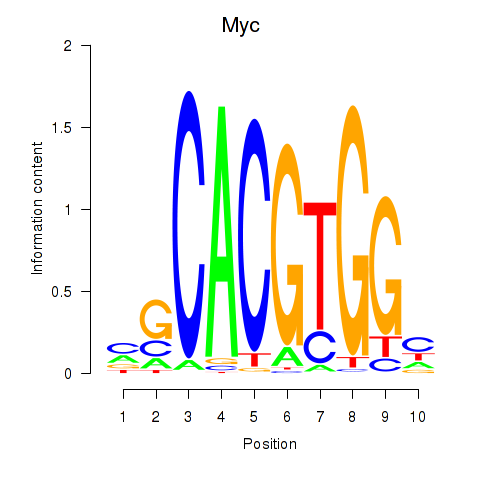
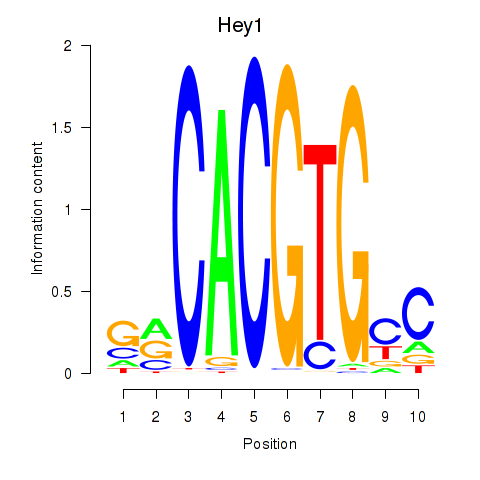
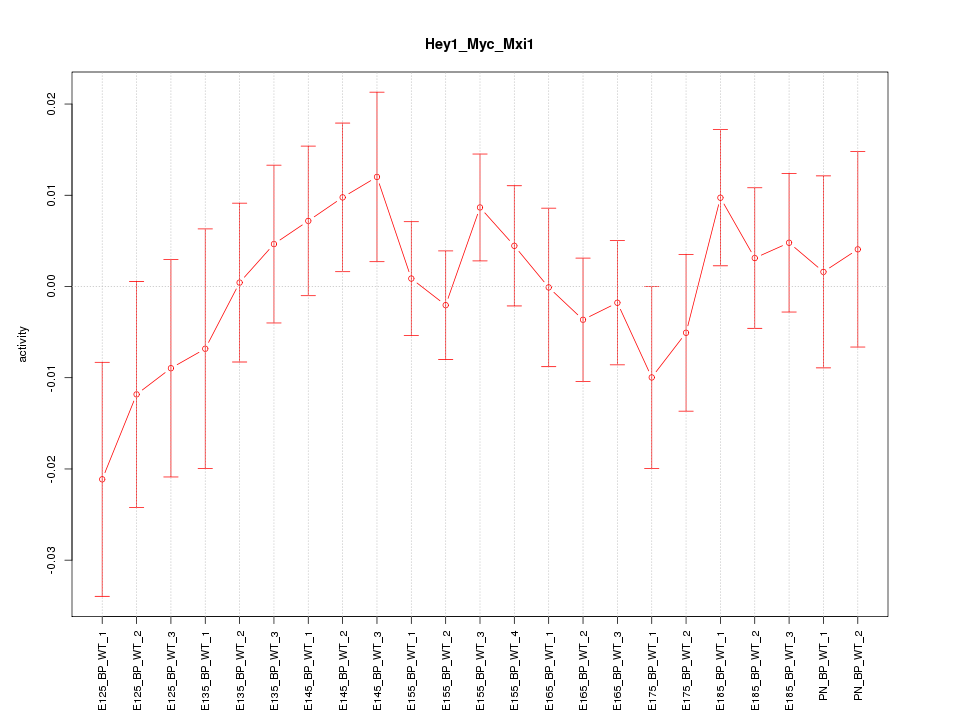
| 1.1 |
8.0 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.8 |
2.5 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.8 |
2.4 |
GO:0046881 |
positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.8 |
2.3 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 0.7 |
2.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.6 |
1.8 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.6 |
2.3 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.6 |
1.7 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.5 |
1.5 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.5 |
1.9 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.5 |
0.5 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.4 |
1.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 |
1.3 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 |
1.2 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 |
1.6 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 |
1.9 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.4 |
1.8 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 |
1.2 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 |
0.9 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 |
1.5 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 |
0.8 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 |
1.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 |
0.8 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.3 |
1.4 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 |
0.8 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.3 |
1.1 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
1.0 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 0.2 |
0.5 |
GO:0034144 |
negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 |
3.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
0.7 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 |
0.5 |
GO:0072197 |
ureter morphogenesis(GO:0072197) |
| 0.2 |
1.4 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.2 |
2.4 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
0.7 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 |
1.2 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.2 |
1.1 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.2 |
0.7 |
GO:0046370 |
fructose biosynthetic process(GO:0046370) |
| 0.2 |
0.7 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.2 |
0.9 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
1.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 |
0.6 |
GO:0090403 |
regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) oxidative stress-induced premature senescence(GO:0090403) negative regulation of fermentation(GO:1901003) |
| 0.2 |
1.5 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.2 |
1.5 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.2 |
0.6 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 |
0.8 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 |
1.2 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.2 |
1.0 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 |
1.3 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.2 |
0.5 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 |
0.5 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 |
0.5 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.2 |
0.9 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 |
0.5 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 |
0.3 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.2 |
0.9 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.2 |
3.6 |
GO:0046033 |
AMP metabolic process(GO:0046033) |
| 0.2 |
0.7 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.2 |
1.8 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.2 |
0.8 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.2 |
0.7 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.2 |
1.8 |
GO:0046051 |
UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 |
1.0 |
GO:0021960 |
anterior commissure morphogenesis(GO:0021960) |
| 0.2 |
0.6 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.2 |
1.1 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 |
0.6 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 |
1.4 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.2 |
1.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 |
0.6 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 |
0.3 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
1.6 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 |
0.3 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.4 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
2.3 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 |
0.6 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.1 |
0.6 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.5 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
0.7 |
GO:0090292 |
nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.8 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 |
1.4 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
2.7 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
1.5 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.6 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 |
0.7 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
1.1 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 |
0.1 |
GO:0045590 |
negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 |
0.4 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.5 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
1.0 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.4 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.1 |
0.4 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
0.8 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
0.2 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.1 |
0.6 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.4 |
GO:0070625 |
zymogen granule exocytosis(GO:0070625) |
| 0.1 |
1.9 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.5 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.2 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.6 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
0.9 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.3 |
GO:0006207 |
'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 |
0.6 |
GO:0015819 |
lysine transport(GO:0015819) |
| 0.1 |
0.9 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.7 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
0.6 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.3 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.8 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
0.7 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
2.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.6 |
GO:0009125 |
nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 |
0.1 |
GO:1901536 |
regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 |
0.2 |
GO:0097252 |
oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 |
0.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.5 |
GO:2000110 |
negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 |
0.7 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.1 |
0.7 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
0.5 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.6 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.4 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 |
0.9 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.4 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
1.1 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.1 |
1.6 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.1 |
0.3 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.7 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.6 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.2 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 |
0.2 |
GO:0050651 |
dermatan sulfate biosynthetic process(GO:0030208) dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 |
0.4 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.4 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.3 |
GO:1903054 |
late endosomal microautophagy(GO:0061738) lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.3 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.1 |
1.1 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.1 |
0.5 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.2 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 |
0.3 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 |
0.3 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.1 |
0.4 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.5 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.5 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.4 |
GO:0021508 |
floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.2 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.3 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
0.3 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.5 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 |
0.2 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
1.1 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.3 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.1 |
1.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.1 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.3 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.1 |
0.1 |
GO:0042362 |
fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 |
0.7 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.4 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.4 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
1.5 |
GO:0051560 |
mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 |
0.1 |
GO:1903689 |
regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 |
0.3 |
GO:1903598 |
negative regulation of pinocytosis(GO:0048550) angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.1 |
0.6 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.2 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
0.3 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
0.3 |
GO:0061101 |
neuroendocrine cell differentiation(GO:0061101) |
| 0.1 |
0.4 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.3 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
0.3 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.4 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.3 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 |
0.4 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
0.2 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.1 |
0.5 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
0.4 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.1 |
0.3 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
1.1 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
0.4 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 |
0.5 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 |
0.2 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.1 |
0.1 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
0.2 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.2 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 |
0.4 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.2 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
0.2 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 |
0.4 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
0.6 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
1.1 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
1.0 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.1 |
0.1 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.1 |
0.7 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) |
| 0.1 |
0.6 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
1.4 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
0.4 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.1 |
0.3 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.2 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 |
0.2 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
0.2 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 |
0.2 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.4 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 |
0.3 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
0.3 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.1 |
GO:1902548 |
negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 |
0.7 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.5 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.4 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 |
0.2 |
GO:0030656 |
regulation of vitamin metabolic process(GO:0030656) |
| 0.0 |
1.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.8 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
1.0 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.1 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 |
0.5 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.4 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.1 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.0 |
0.1 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.1 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation(GO:0018307) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 |
1.0 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 |
0.1 |
GO:0035826 |
rubidium ion transport(GO:0035826) |
| 0.0 |
1.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.6 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.3 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 |
0.1 |
GO:1903750 |
regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 |
0.3 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.9 |
GO:0033173 |
calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 |
0.6 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.6 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.4 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.9 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 |
0.5 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.3 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.7 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.2 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.1 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.0 |
0.1 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.0 |
GO:1900368 |
regulation of RNA interference(GO:1900368) |
| 0.0 |
0.2 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.1 |
GO:0021798 |
forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 |
0.6 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.5 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.2 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.6 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.0 |
0.3 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.1 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 |
0.2 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.2 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 |
0.4 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.1 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 |
1.0 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.2 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.0 |
0.1 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
0.3 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
0.2 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.6 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.0 |
0.1 |
GO:0060066 |
oviduct development(GO:0060066) |
| 0.0 |
0.1 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 |
0.1 |
GO:0003256 |
regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.0 |
0.3 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 |
0.7 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
1.4 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.1 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.5 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.2 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 |
0.1 |
GO:0019659 |
fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.1 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.5 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.8 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.2 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.7 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.1 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.2 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.1 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.0 |
1.8 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.1 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.2 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.1 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.0 |
0.2 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.7 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
0.1 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.2 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.0 |
0.1 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.2 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.4 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.7 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.2 |
GO:0048701 |
embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 |
0.1 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.2 |
GO:0010717 |
regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.4 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 |
1.3 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
0.1 |
GO:0034134 |
toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 |
0.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
0.1 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.3 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.4 |
GO:0010470 |
regulation of gastrulation(GO:0010470) |
| 0.0 |
0.6 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.2 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.1 |
GO:0006145 |
purine nucleobase catabolic process(GO:0006145) |
| 0.0 |
0.5 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.2 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.2 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 |
0.2 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.2 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.3 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.0 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.4 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.3 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.0 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.1 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.5 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 |
0.7 |
GO:0099601 |
regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 |
0.1 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.5 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.3 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.0 |
0.5 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.1 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.9 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.5 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.2 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.0 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 |
0.4 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.1 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.5 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.2 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 |
0.1 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.1 |
GO:0071880 |
adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 |
0.2 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.2 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 |
0.2 |
GO:0051091 |
positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 |
0.5 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.4 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.2 |
GO:0003338 |
metanephros morphogenesis(GO:0003338) |
| 0.0 |
0.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.2 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.9 |
GO:0046330 |
positive regulation of JNK cascade(GO:0046330) |
| 0.0 |
0.1 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.0 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.0 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.2 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.0 |
GO:0019471 |
4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 |
0.1 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
1.1 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.0 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.2 |
GO:0006919 |
activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 |
0.1 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.1 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.5 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.3 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.0 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 |
0.2 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.2 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.2 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.1 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.1 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.1 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.1 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.0 |
0.1 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.0 |
GO:1990000 |
amyloid fibril formation(GO:1990000) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.0 |
GO:2000772 |
regulation of cellular senescence(GO:2000772) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.1 |
GO:0045002 |
double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.3 |
GO:0071391 |
cellular response to estrogen stimulus(GO:0071391) |
| 0.0 |
0.0 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.0 |
0.3 |
GO:0007044 |
cell-substrate junction assembly(GO:0007044) |
| 0.0 |
0.3 |
GO:0050850 |
positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 |
0.0 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.0 |
0.6 |
GO:0030641 |
regulation of cellular pH(GO:0030641) |
| 0.0 |
0.1 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.0 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.1 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.1 |
GO:0070970 |
interleukin-2 secretion(GO:0070970) regulation of interleukin-2 secretion(GO:1900040) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.9 |
GO:0000187 |
activation of MAPK activity(GO:0000187) |
| 0.0 |
0.0 |
GO:0034766 |
negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 |
0.2 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.8 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.0 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.1 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.0 |
0.0 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.3 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.1 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
0.0 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.2 |
GO:0090305 |
nucleic acid phosphodiester bond hydrolysis(GO:0090305) |