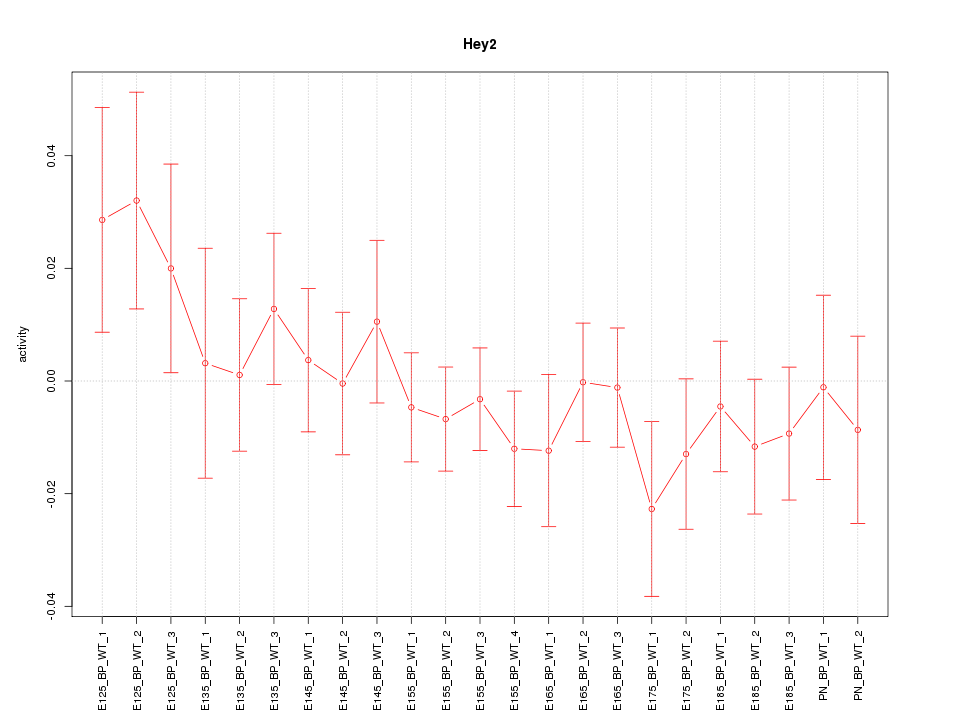
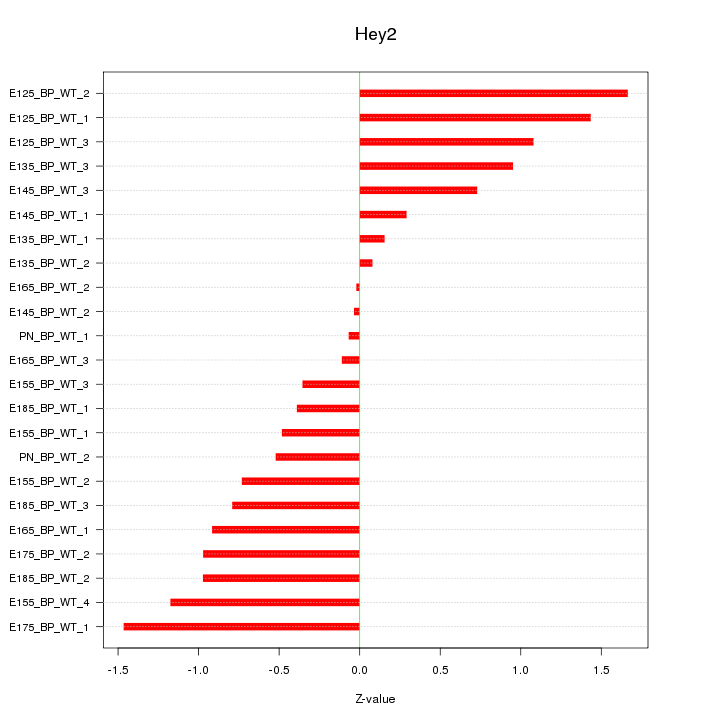
Motif ID: Hey2
Z-value: 0.828
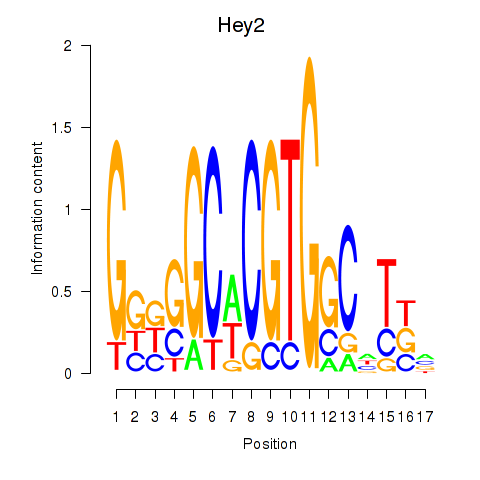
Transcription factors associated with Hey2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hey2 | ENSMUSG00000019789.8 | Hey2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | mm10_v2_chr10_-_30842765_30842801 | -0.61 | 2.0e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0071205 | clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 | 7.0 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.5 | 1.9 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.4 | 0.9 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.3 | 2.8 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.3 | 0.9 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.3 | 0.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 0.7 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 | 0.9 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 0.8 | GO:0031622 | positive regulation of fever generation(GO:0031622) |
| 0.2 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 0.7 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.6 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 1.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 5.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.4 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.5 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 1.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 1.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0019405 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.1 | 1.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.5 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.7 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.7 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 1.2 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) positive regulation of keratinocyte differentiation(GO:0045618) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.6 | GO:0030811 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) |
| 0.0 | 1.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.5 | GO:1900087 | ventricular system development(GO:0021591) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 3.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.9 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 2.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 3.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 5.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 4.7 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.3 | 2.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 7.0 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.2 | 1.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 0.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 2.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 5.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |