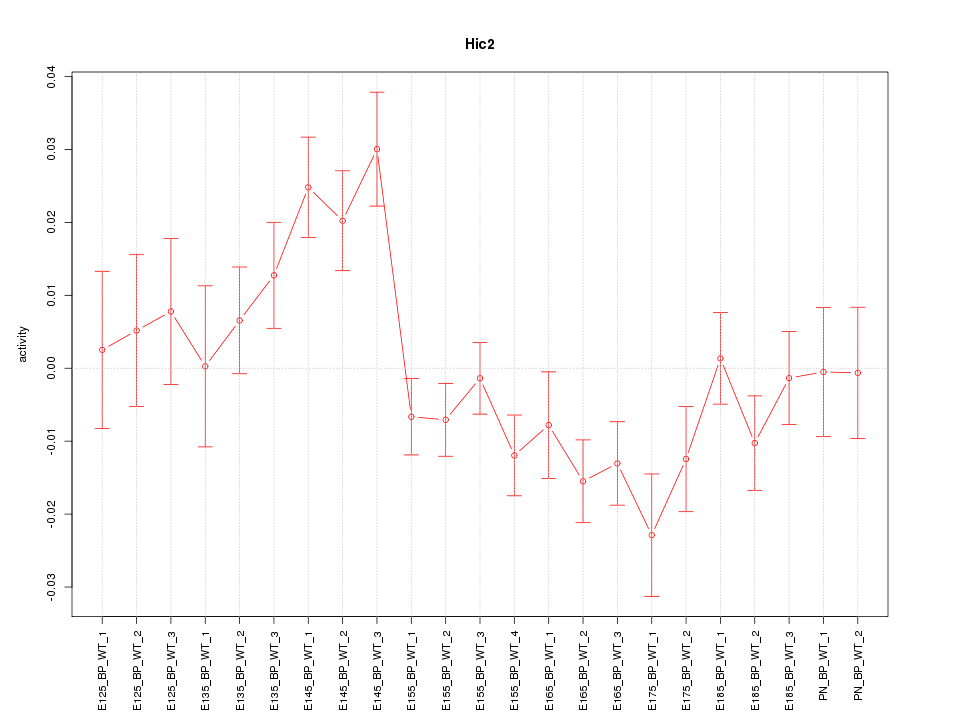
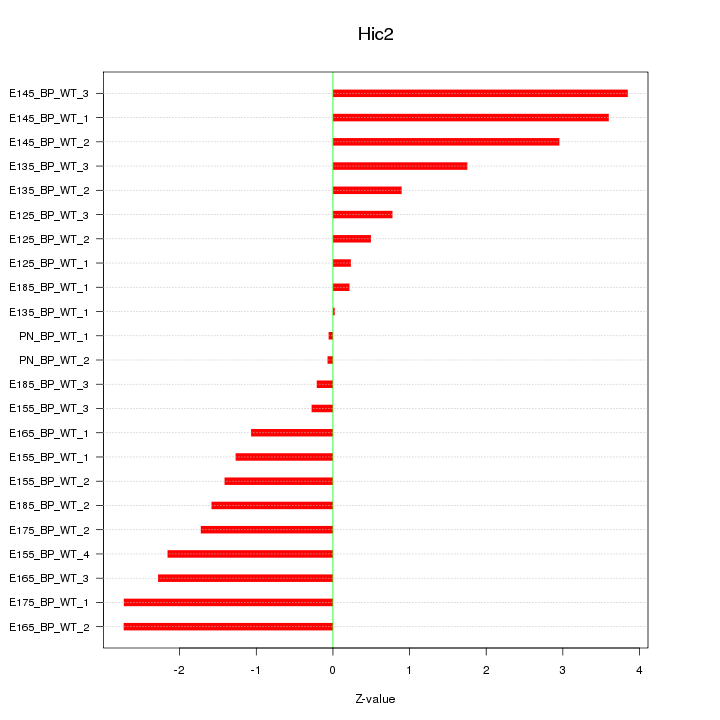
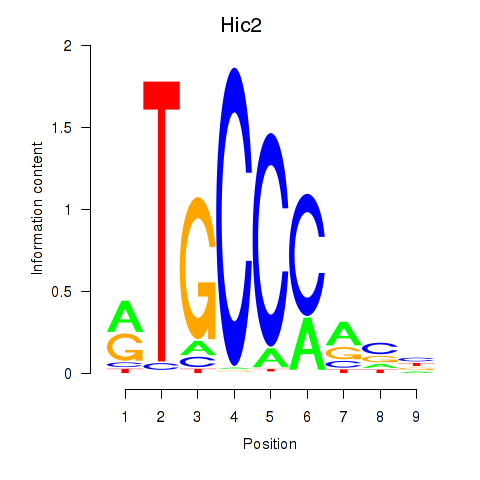
Motif ID: Hic2
Z-value: 1.823
Transcription factors associated with Hic2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hic2 | ENSMUSG00000050240.8 | Hic2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic2 | mm10_v2_chr16_+_17233560_17233664 | -0.14 | 5.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.5 | 4.6 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 1.2 | 3.6 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 1.1 | 4.3 | GO:0003360 | brainstem development(GO:0003360) |
| 1.0 | 5.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.0 | 3.1 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.8 | 10.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.7 | 2.2 | GO:0032305 | positive regulation of icosanoid secretion(GO:0032305) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.7 | 2.6 | GO:0021586 | pons maturation(GO:0021586) |
| 0.6 | 3.9 | GO:0032796 | uropod organization(GO:0032796) |
| 0.6 | 1.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.6 | 1.9 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.6 | 2.3 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.6 | 3.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.6 | 2.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.6 | 1.7 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.6 | 2.8 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.5 | 2.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.5 | 1.6 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.5 | 2.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 2.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 0.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.5 | 2.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.5 | 0.5 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.5 | 1.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.4 | 1.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 0.4 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.4 | 1.2 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.2 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.4 | 1.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 1.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 2.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.4 | 4.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 1.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 1.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) trachea cartilage morphogenesis(GO:0060535) |
| 0.4 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.4 | 1.8 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.3 | 1.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 1.7 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.3 | 1.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 5.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 11.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 1.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.3 | 2.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.9 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 2.2 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 2.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 1.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 0.9 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 1.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 7.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 2.0 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.3 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 0.8 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 0.8 | GO:1902219 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 0.6 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.3 | 1.9 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.3 | 0.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 1.1 | GO:1903984 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 0.5 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.3 | 3.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 0.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.2 | 1.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.2 | 1.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.2 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 2.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 0.5 | GO:0001828 | inner cell mass cell differentiation(GO:0001826) inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 | 0.7 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.2 | 0.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 1.6 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 1.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) response to glucagon(GO:0033762) |
| 0.2 | 1.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 2.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 3.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 1.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 1.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 2.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 0.8 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.2 | 0.6 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.2 | 1.4 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 3.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 0.4 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.2 | 0.4 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.2 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.6 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 3.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 2.0 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 2.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 3.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 1.8 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.2 | 1.7 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.2 | 0.7 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.2 | 0.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 1.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.5 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.2 | 0.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 0.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.8 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 1.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 0.8 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.6 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 4.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.4 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 2.3 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 1.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.3 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.1 | 2.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 1.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0048866 | stem cell fate specification(GO:0048866) |
| 0.1 | 0.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 2.6 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 1.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 1.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 2.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:0090529 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.4 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 1.6 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.1 | 0.6 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.7 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 3.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.5 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.8 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.2 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.7 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.4 | GO:0071459 | kinetochore assembly(GO:0051382) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.5 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.1 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 1.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 4.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.9 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.5 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.9 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.8 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 1.0 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 1.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 0.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.4 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.6 | GO:0019348 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.1 | 0.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.5 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.3 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 1.9 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 3.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.3 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 0.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.9 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.1 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.3 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 2.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.3 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.7 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 1.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:2001197 | regulation of extracellular matrix assembly(GO:1901201) positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 1.9 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0072683 | T cell extravasation(GO:0072683) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 2.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.6 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0032402 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.9 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.4 | GO:0014741 | negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0019755 | one-carbon compound transport(GO:0019755) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.4 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.4 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.4 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.9 | 2.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.8 | 12.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 3.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 2.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 1.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 4.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 2.6 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 1.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 3.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 4.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 0.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 3.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 3.0 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.2 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 1.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 3.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 4.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.5 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.1 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 4.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.8 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 9.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.8 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 3.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 8.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.3 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 3.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.9 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) proteasome accessory complex(GO:0022624) |
| 0.0 | 1.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 1.2 | 3.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 1.0 | 12.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 4.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.7 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 2.6 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.7 | 2.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 4.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 3.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 3.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.6 | 2.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.6 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) |
| 0.5 | 2.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.5 | 1.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.5 | 1.4 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.4 | 1.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.4 | 1.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 0.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.6 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.4 | 3.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 1.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.3 | 2.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 2.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 1.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 1.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 1.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 1.7 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 0.8 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 1.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.3 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 1.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 1.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.7 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.5 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.2 | 0.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 5.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.5 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 4.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 3.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.2 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 1.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 3.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.5 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.2 | 1.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 7.8 | GO:0001653 | peptide receptor activity(GO:0001653) |
| 0.2 | 3.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.4 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.1 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 3.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 1.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.1 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.4 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.4 | GO:0001851 | complement component C3b binding(GO:0001851) proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 4.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.4 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 3.5 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.1 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 2.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0044388 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) small protein activating enzyme binding(GO:0044388) cupric ion binding(GO:1903135) cuprous ion binding(GO:1903136) glyoxalase (glycolic acid-forming) activity(GO:1990422) |
| 0.1 | 0.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.1 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 1.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.6 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.0 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 3.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 1.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0030362 | protein phosphatase type 4 regulator activity(GO:0030362) |
| 0.0 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.1 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |