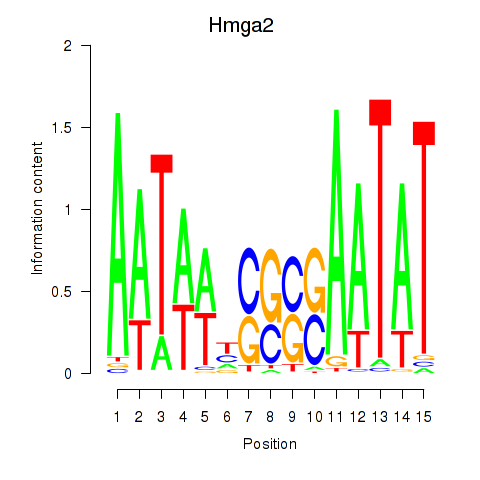
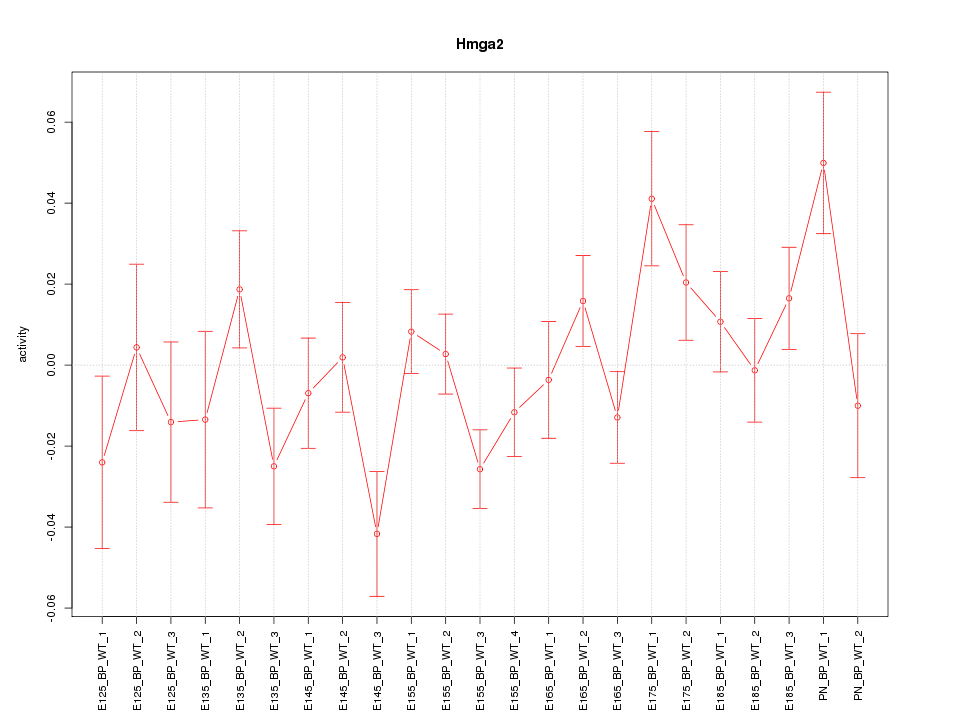
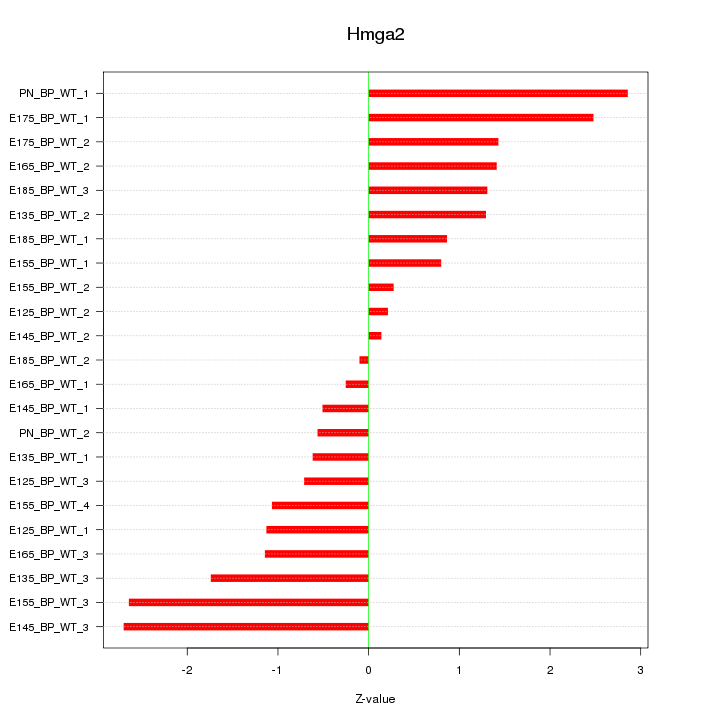
| 1.3 |
7.9 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.6 |
1.8 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 |
2.4 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
0.7 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) positive regulation of cortisol secretion(GO:0051464) |
| 0.2 |
1.0 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 |
0.8 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 |
0.5 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 |
1.2 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 |
1.5 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
1.0 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 |
1.8 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.1 |
1.2 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
2.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
1.7 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 |
3.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.5 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.3 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
1.0 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.3 |
GO:2000561 |
CD4-positive, alpha-beta T cell proliferation(GO:0035739) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 |
0.3 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.9 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 |
0.4 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
0.5 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
0.2 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
0.5 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
0.4 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 |
0.3 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
0.8 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.6 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.1 |
0.4 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.1 |
1.1 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 |
1.0 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.6 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.0 |
0.6 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.8 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
1.1 |
GO:0006862 |
nucleotide transport(GO:0006862) |
| 0.0 |
0.8 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.2 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.6 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.3 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.7 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.1 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.0 |
0.1 |
GO:2000427 |
regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 |
1.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.3 |
GO:0000478 |
endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 |
0.8 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
1.6 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
1.4 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.3 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) |
| 0.0 |
0.2 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.4 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.3 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.4 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.3 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |