Motif ID: Hmx1
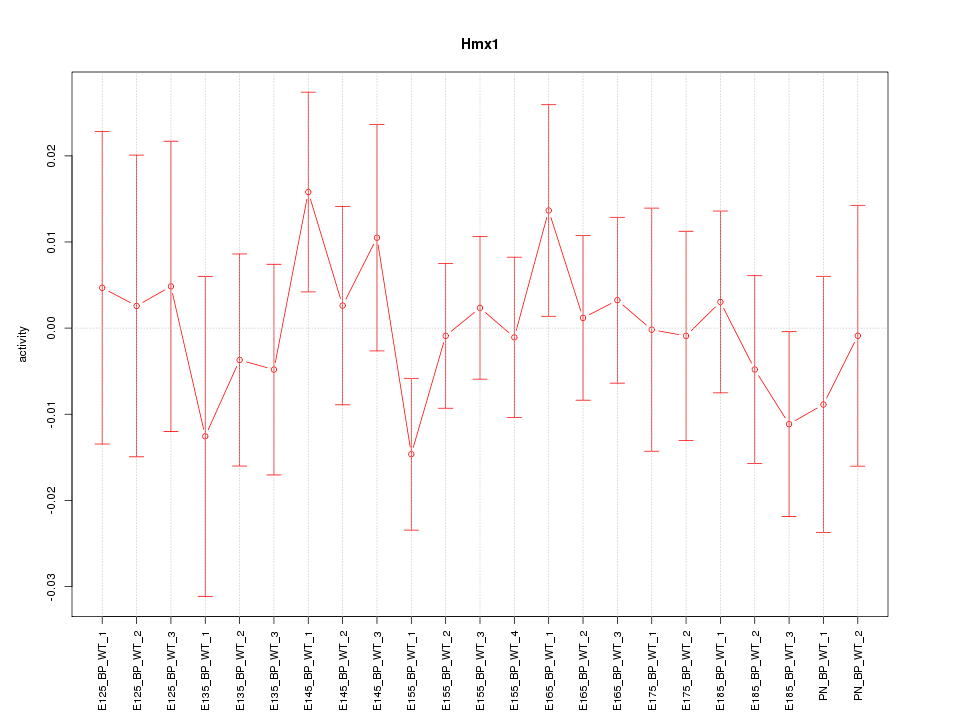
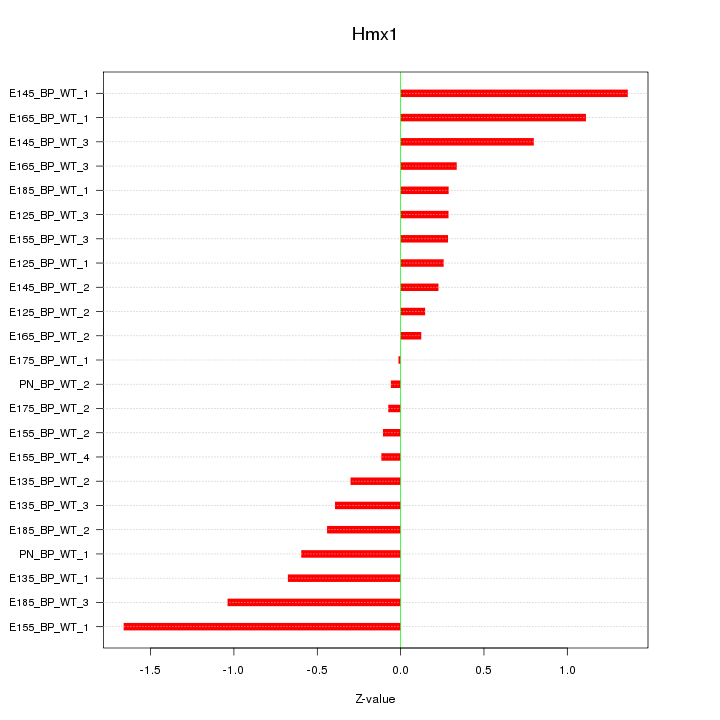
Z-value: 0.638
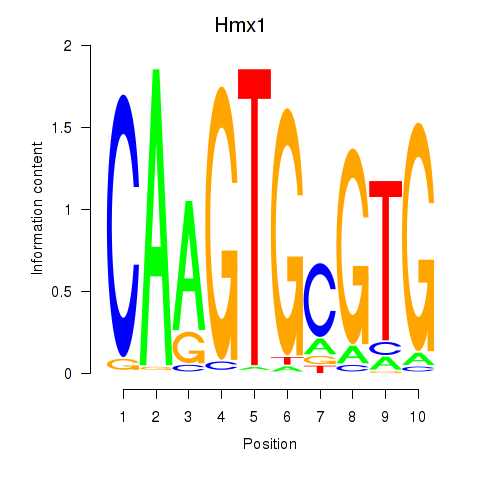
Transcription factors associated with Hmx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hmx1 | ENSMUSG00000067438.3 | Hmx1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0072070 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:1901524 | regulation of autophagosome maturation(GO:1901096) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.2 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:2001137 | protein retention in Golgi apparatus(GO:0045053) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) regulation of cholesterol import(GO:0060620) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.1 | GO:0098543 | detection of bacterium(GO:0016045) negative regulation of natural killer cell activation(GO:0032815) detection of other organism(GO:0098543) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.5 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0034702 | ion channel complex(GO:0034702) transmembrane transporter complex(GO:1902495) transporter complex(GO:1990351) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.2 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |