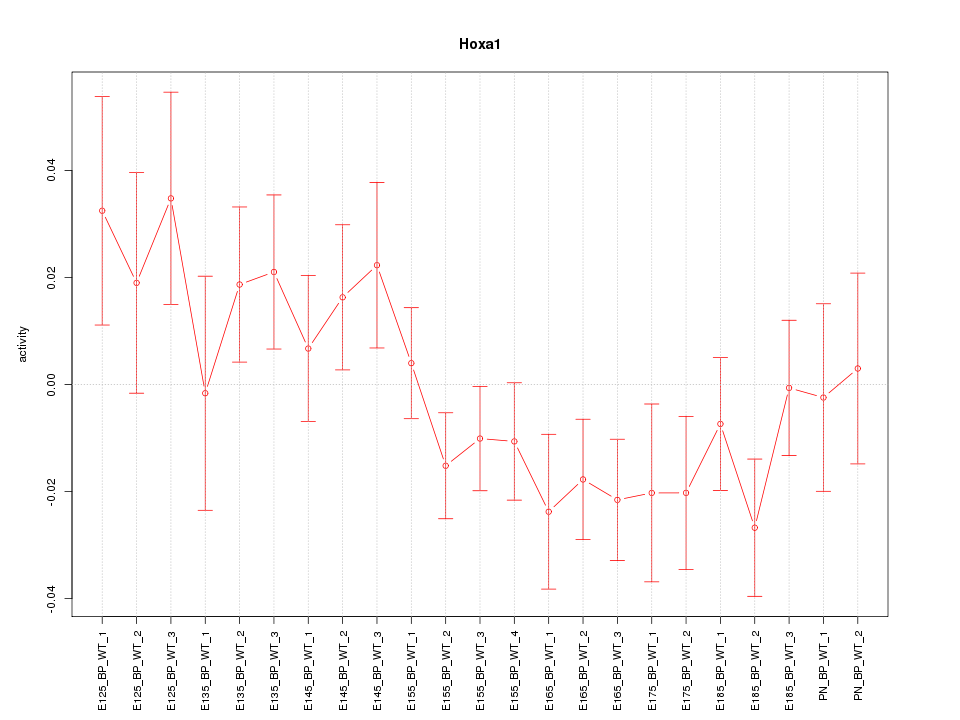
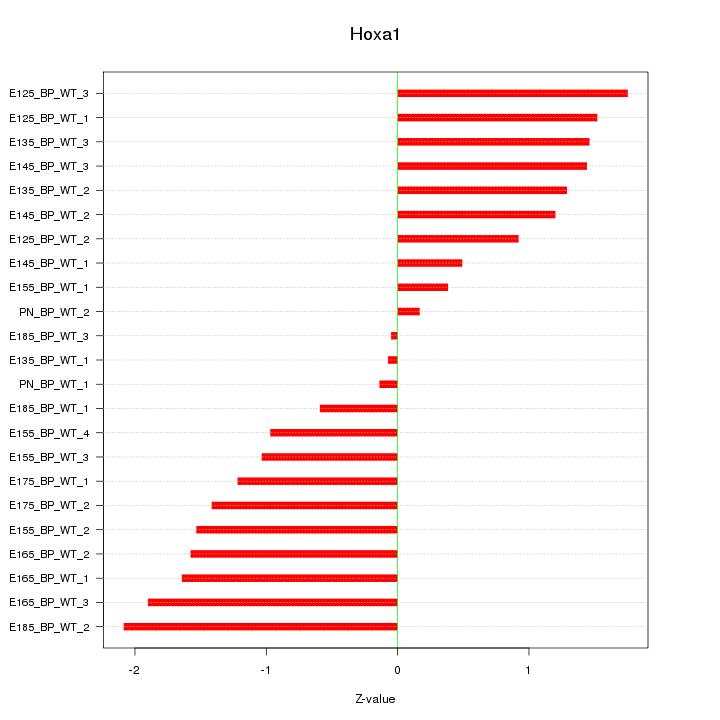
Motif ID: Hoxa1
Z-value: 1.240
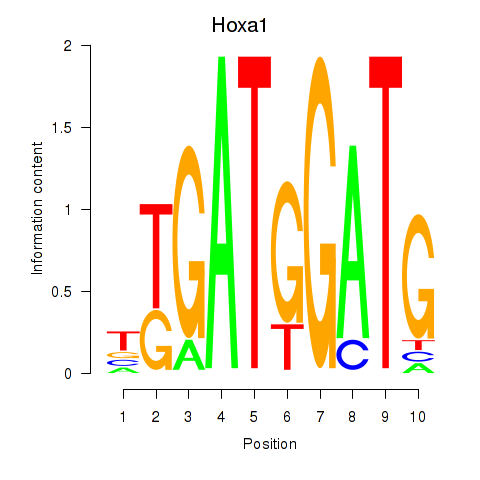
Transcription factors associated with Hoxa1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa1 | ENSMUSG00000029844.9 | Hoxa1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.7 | 4.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 6.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.4 | 4.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 2.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 3.6 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 3.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 2.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 0.9 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 1.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 1.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 2.3 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 3.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 2.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 4.3 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 1.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 6.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.6 | GO:0008517 | folic acid transporter activity(GO:0008517) water channel activity(GO:0015250) |
| 0.6 | 4.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 4.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 3.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.7 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 6.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 3.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |