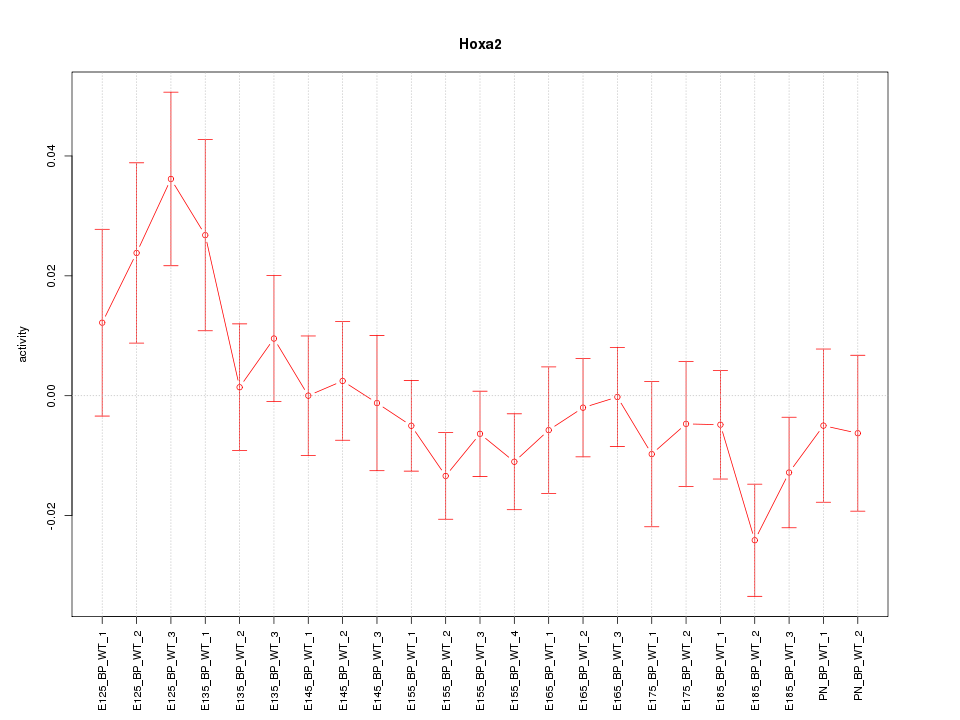
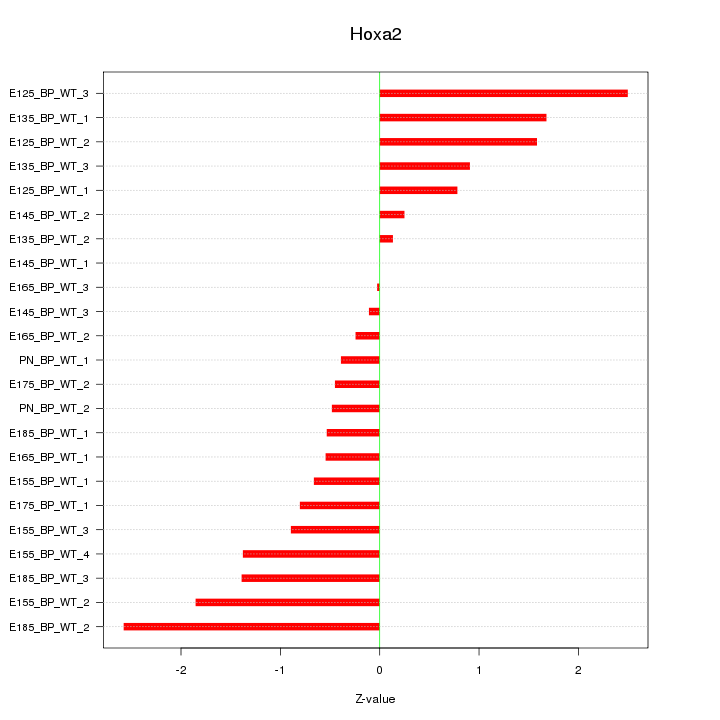
Motif ID: Hoxa2
Z-value: 1.144
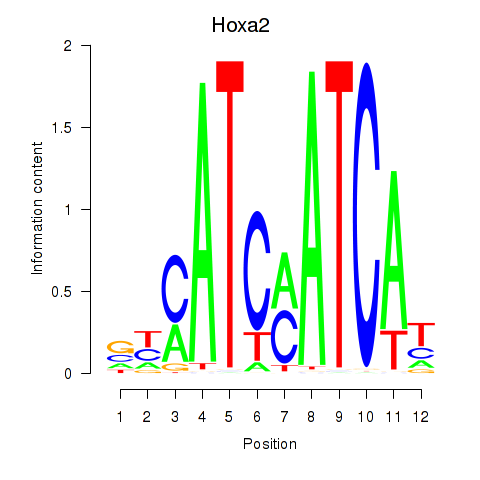
Transcription factors associated with Hoxa2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa2 | ENSMUSG00000014704.8 | Hoxa2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 19.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.5 | 10.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.2 | 3.5 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.7 | 2.1 | GO:1902713 | regulation of interferon-gamma secretion(GO:1902713) |
| 0.5 | 1.6 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.5 | 0.5 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.5 | 1.4 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.5 | 1.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.4 | 1.8 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.4 | 2.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 1.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 2.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 1.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 2.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 0.6 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.3 | 0.6 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.2 | 1.2 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.2 | 1.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 2.8 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 1.6 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 0.7 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 3.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 0.7 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 | 2.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 5.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.8 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 1.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 0.7 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 0.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.5 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.4 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.3 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.5 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 1.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.4 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) motor learning(GO:0061743) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.6 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.4 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.1 | 0.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 1.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.3 | GO:0072610 | interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.7 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.2 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 4.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.6 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.2 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.7 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.1 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.2 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.2 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 3.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 2.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 3.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.8 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 1.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.5 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.7 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 1.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.5 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 1.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.7 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 2.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.4 | 1.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.3 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 2.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 8.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 3.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 4.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 13.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 22.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.2 | 3.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.5 | 2.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 2.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 2.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 1.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.7 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 3.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 5.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 2.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.5 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 2.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0047598 | 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.6 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 4.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.4 | GO:0023026 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.7 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 1.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 2.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.9 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.9 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.1 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) sterol O-acyltransferase activity(GO:0004772) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) protein-cysteine S-acyltransferase activity(GO:0019707) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) Ras palmitoyltransferase activity(GO:0043849) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.6 | GO:0043178 | alcohol binding(GO:0043178) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 2.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0023029 | peptide antigen-transporting ATPase activity(GO:0015433) MHC class Ib protein binding(GO:0023029) tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |