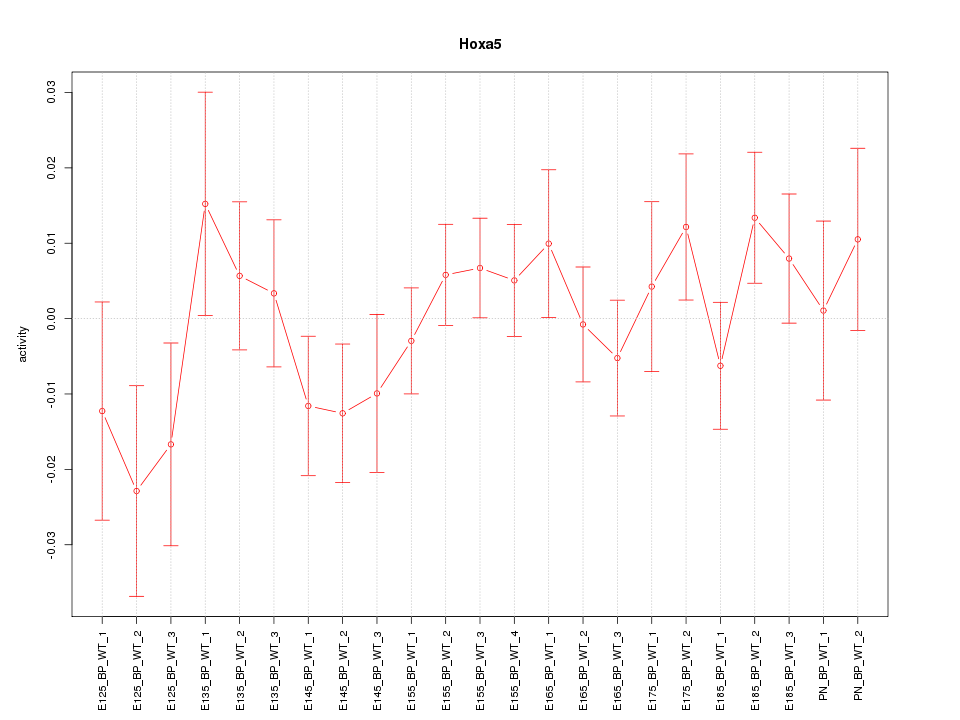
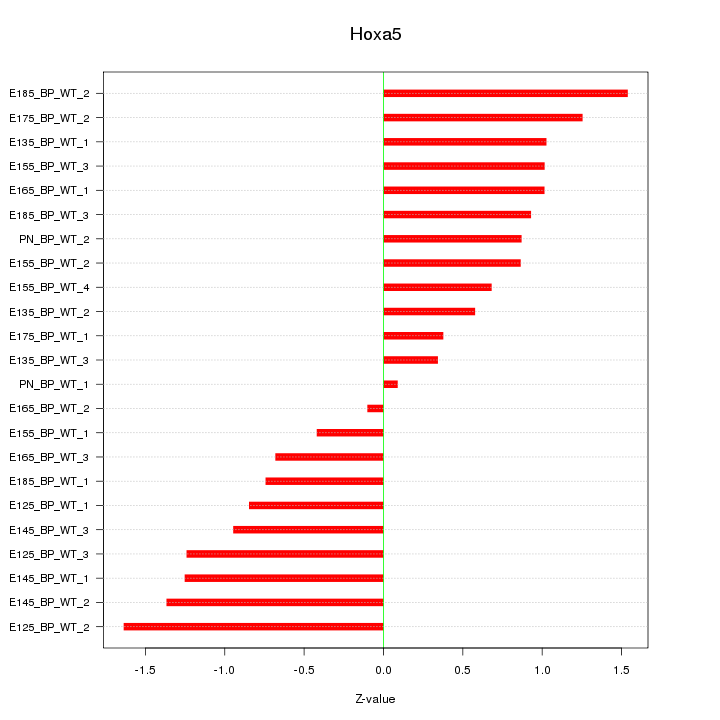
Motif ID: Hoxa5
Z-value: 0.955
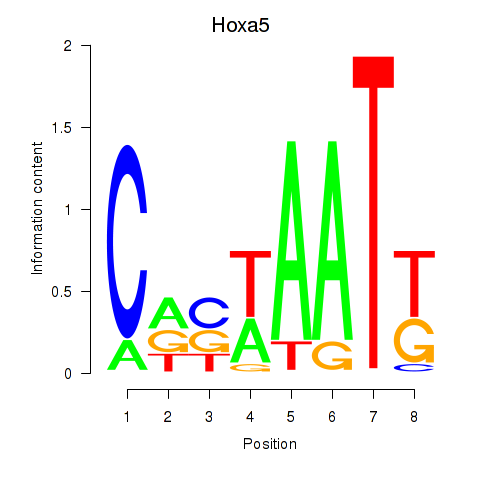
Transcription factors associated with Hoxa5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa5 | ENSMUSG00000038253.6 | Hoxa5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.6 | 1.9 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 | 1.0 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 | 1.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.3 | 1.0 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 | 1.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 0.8 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.2 | 1.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.6 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 | 0.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 0.6 | GO:0048866 | stem cell fate specification(GO:0048866) |
| 0.2 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.2 | 0.2 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 | 0.5 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 0.6 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.2 | 0.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 2.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.7 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 4.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.4 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 1.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.4 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 | 0.5 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.7 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.7 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.3 | GO:2001180 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 0.6 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.4 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.1 | GO:0007492 | endoderm formation(GO:0001706) endoderm development(GO:0007492) endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 0.6 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.1 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.5 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 1.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 2.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.2 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.3 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) positive regulation of mast cell chemotaxis(GO:0060754) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 1.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 2.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0032229 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 2.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 2.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.5 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.4 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.5 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.2 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.2 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0046541 | regulation of vascular smooth muscle contraction(GO:0003056) saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0035880 | cell proliferation in midbrain(GO:0033278) embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:2000807 | synaptic vesicle clustering(GO:0097091) regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.0 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0000237 | leptotene(GO:0000237) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:0045191 | regulation of isotype switching(GO:0045191) |
| 0.0 | 0.3 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.4 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.8 | 8.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 1.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.3 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.3 | 1.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 0.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 1.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0032450 | oligo-1,6-glucosidase activity(GO:0004574) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 1.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.1 | 0.6 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 2.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.3 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 0.6 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 5.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.4 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.0 | GO:0044388 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) small protein activating enzyme binding(GO:0044388) cupric ion binding(GO:1903135) cuprous ion binding(GO:1903136) glyoxalase (glycolic acid-forming) activity(GO:1990422) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.7 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.9 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |