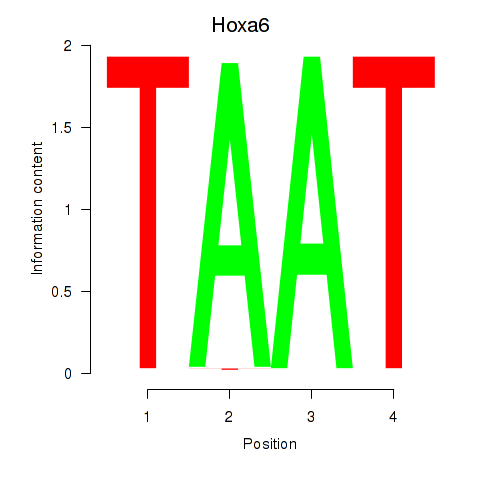
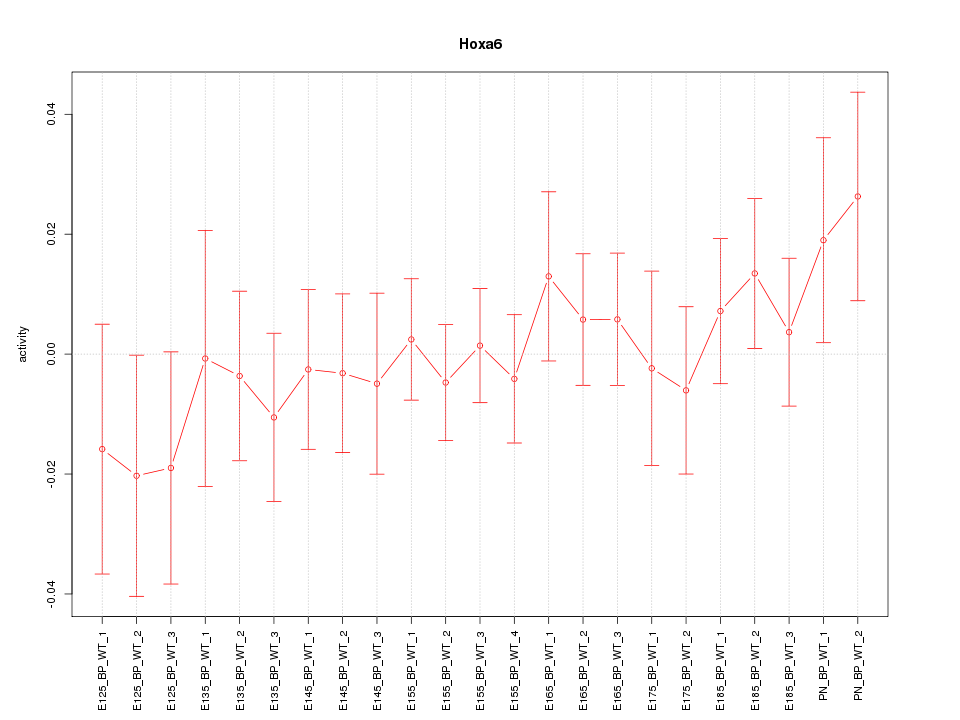
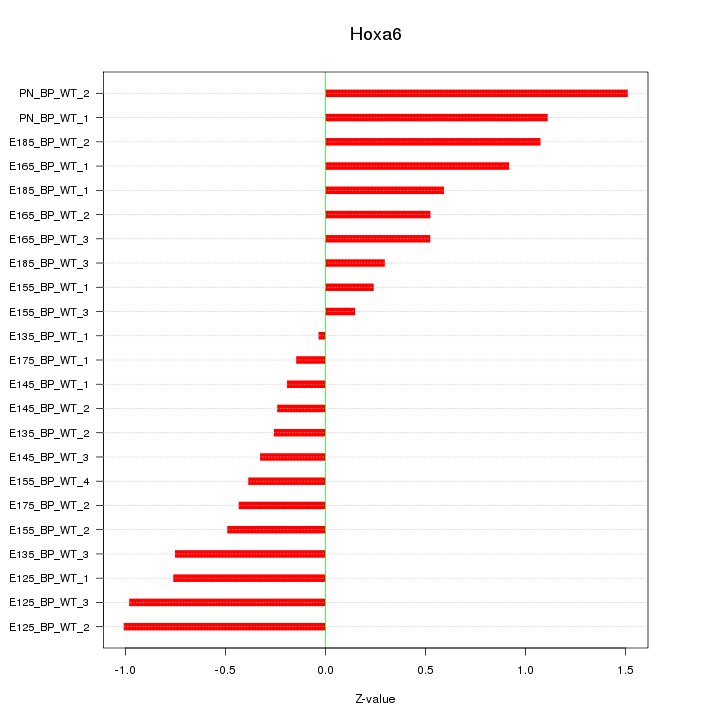
| 2.0 |
5.9 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.1 |
4.5 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 1.0 |
6.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.9 |
3.5 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.7 |
6.7 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.7 |
2.7 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.6 |
3.6 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.5 |
1.0 |
GO:0072095 |
regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.5 |
1.4 |
GO:1900020 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.4 |
14.1 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.4 |
2.3 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.4 |
2.5 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 |
4.4 |
GO:1901629 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of presynaptic membrane organization(GO:1901629) |
| 0.3 |
1.4 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.3 |
3.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
0.5 |
GO:0061324 |
canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.2 |
0.7 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 |
0.9 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 |
0.8 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.2 |
0.6 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 |
1.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
0.5 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.2 |
0.7 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 |
1.6 |
GO:0021960 |
anterior commissure morphogenesis(GO:0021960) |
| 0.2 |
1.6 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
1.7 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.2 |
2.3 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.2 |
6.1 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.2 |
1.5 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
1.0 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
2.9 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.1 |
1.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
1.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.5 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
2.4 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
0.3 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 |
2.1 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.3 |
GO:1990314 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 |
0.5 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 |
1.1 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.1 |
0.9 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
5.5 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.1 |
0.4 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 |
0.5 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.1 |
1.3 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
2.9 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 |
0.1 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 |
3.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.7 |
GO:0035584 |
calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 |
0.2 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
1.1 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
1.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
1.0 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.4 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.4 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.3 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.4 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.8 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
4.7 |
GO:0007160 |
cell-matrix adhesion(GO:0007160) |
| 0.0 |
0.1 |
GO:0097107 |
postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.9 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
1.0 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.4 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
1.2 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.5 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.1 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.0 |
2.0 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.3 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.8 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |