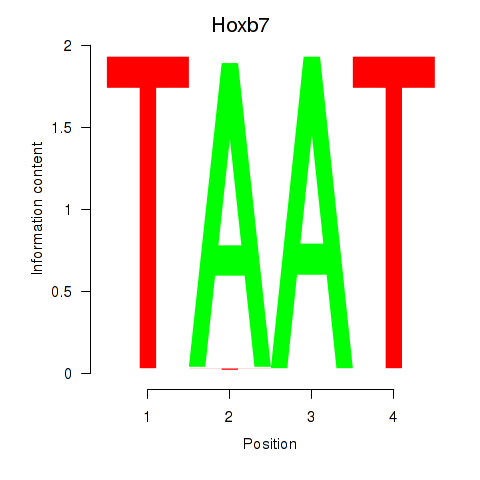
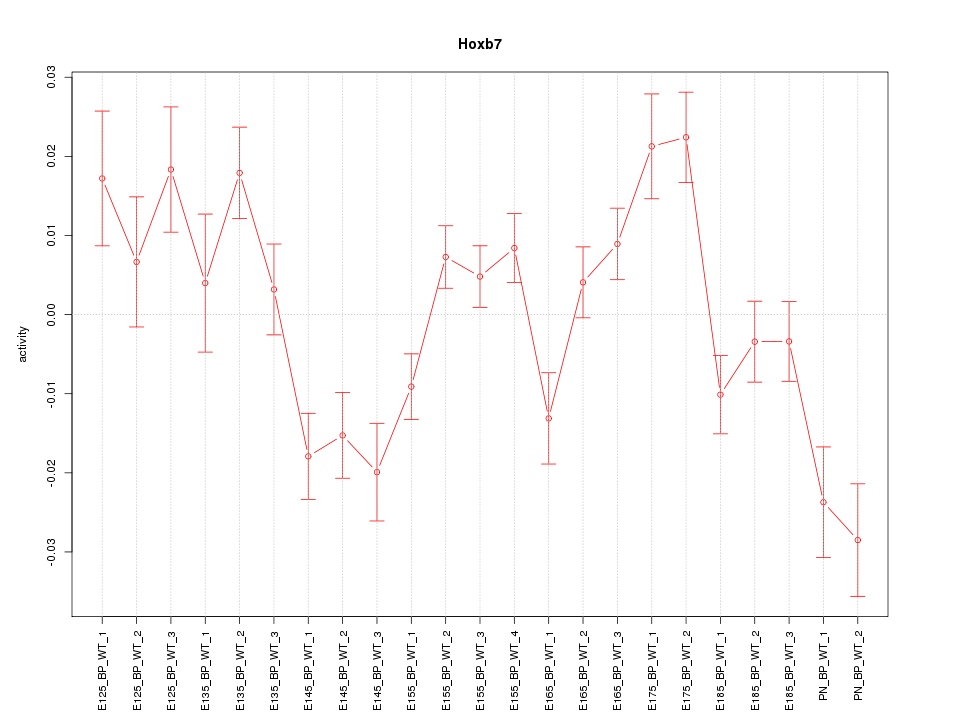
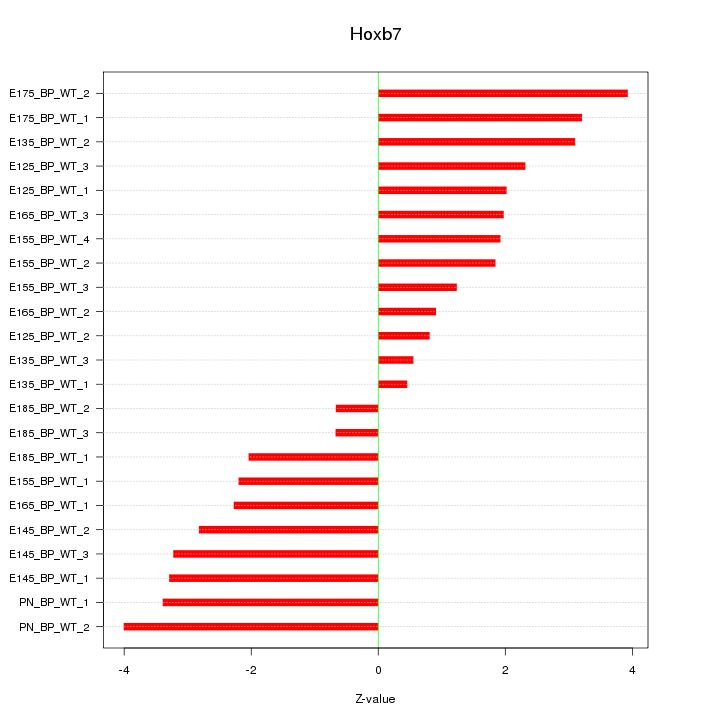
| 3.1 |
9.4 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 2.9 |
8.7 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 2.5 |
7.4 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 1.6 |
4.8 |
GO:0060853 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 1.4 |
5.7 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 1.3 |
11.7 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.3 |
3.9 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.2 |
3.7 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 1.2 |
13.2 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.1 |
5.7 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 1.1 |
3.4 |
GO:0060023 |
soft palate development(GO:0060023) |
| 1.1 |
15.9 |
GO:0060013 |
righting reflex(GO:0060013) |
| 1.1 |
6.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.1 |
8.9 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 1.1 |
5.5 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 1.0 |
3.0 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.9 |
0.9 |
GO:1901295 |
regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.9 |
3.6 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.9 |
4.5 |
GO:0061642 |
chemoattraction of axon(GO:0061642) |
| 0.9 |
4.5 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.9 |
2.6 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.9 |
2.6 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.8 |
5.0 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.8 |
4.2 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.8 |
2.4 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.8 |
4.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.7 |
2.1 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.7 |
0.7 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.6 |
1.9 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.6 |
0.6 |
GO:0098597 |
observational learning(GO:0098597) |
| 0.6 |
0.6 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.6 |
5.0 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.6 |
5.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.6 |
6.5 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.6 |
0.6 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.6 |
5.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.6 |
4.6 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.6 |
1.7 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.6 |
2.3 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.6 |
2.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.6 |
1.7 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 |
2.2 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.5 |
9.2 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.5 |
22.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.5 |
1.5 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.5 |
2.4 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.5 |
3.9 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.5 |
1.4 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.5 |
3.3 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.5 |
3.8 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.5 |
1.4 |
GO:0046533 |
negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.5 |
1.8 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.5 |
10.4 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.4 |
1.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 |
1.8 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.4 |
1.8 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.4 |
0.9 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.4 |
0.9 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.4 |
1.3 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.4 |
1.3 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.4 |
0.4 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.4 |
1.3 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.4 |
2.1 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.4 |
2.9 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.4 |
1.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.4 |
2.5 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 |
1.2 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 |
2.9 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.4 |
5.3 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.4 |
1.2 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.4 |
1.2 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.4 |
4.7 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 |
3.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 |
2.0 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.4 |
1.2 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 |
1.2 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.4 |
1.2 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 0.4 |
3.1 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 |
1.5 |
GO:0071372 |
response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 |
2.3 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 |
2.3 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
1.5 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 |
0.4 |
GO:0097324 |
melanocyte migration(GO:0097324) |
| 0.4 |
1.1 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 |
8.4 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.4 |
1.8 |
GO:0072015 |
ciliary body morphogenesis(GO:0061073) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.4 |
1.1 |
GO:0050929 |
corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.4 |
3.9 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 |
1.1 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.4 |
4.6 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.4 |
3.9 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 |
1.0 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 |
1.4 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.3 |
2.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.3 |
1.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.3 |
0.7 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 |
3.4 |
GO:0032230 |
positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.3 |
3.6 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 |
2.6 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.3 |
2.6 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.3 |
2.5 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 |
3.8 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.3 |
5.9 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.3 |
0.9 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 |
7.6 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.3 |
1.2 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.3 |
0.9 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 |
0.9 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 |
2.6 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 |
0.6 |
GO:1903243 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.3 |
1.4 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 |
3.6 |
GO:0061339 |
establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.3 |
1.6 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 |
0.8 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.3 |
1.5 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.3 |
1.3 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.2 |
3.2 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.2 |
1.2 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 |
7.6 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 |
0.7 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 |
1.0 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.2 |
0.5 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 |
1.7 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 |
1.4 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
0.5 |
GO:0060982 |
coronary artery morphogenesis(GO:0060982) |
| 0.2 |
1.0 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) negative regulation of action potential(GO:0045759) |
| 0.2 |
5.0 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 |
0.7 |
GO:0071205 |
protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 |
1.9 |
GO:1903421 |
regulation of synaptic vesicle recycling(GO:1903421) |
| 0.2 |
1.7 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.2 |
0.9 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.2 |
0.7 |
GO:0050923 |
regulation of negative chemotaxis(GO:0050923) |
| 0.2 |
4.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.2 |
1.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.2 |
1.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
7.8 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.2 |
1.1 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 |
0.9 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.2 |
7.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.2 |
0.7 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.2 |
0.7 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 |
3.5 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 |
2.0 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
5.5 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 |
1.3 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.2 |
2.5 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.2 |
1.7 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 |
2.3 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.2 |
1.0 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 |
0.8 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.2 |
1.0 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) regulation of cortisol biosynthetic process(GO:2000064) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.2 |
0.6 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.2 |
1.0 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.2 |
0.6 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 |
4.9 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 |
1.6 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 |
1.0 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 |
2.7 |
GO:0048875 |
chemical homeostasis within a tissue(GO:0048875) |
| 0.2 |
1.2 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 |
1.2 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 |
0.6 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.2 |
1.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 |
0.6 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 |
3.4 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 |
3.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 |
1.3 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) |
| 0.2 |
1.7 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 |
0.7 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.2 |
14.4 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.2 |
4.4 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.2 |
0.9 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.2 |
0.9 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
1.3 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 |
0.5 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 |
0.9 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 |
8.1 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 |
0.5 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.2 |
0.7 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 |
0.3 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.2 |
2.9 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 |
0.8 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 |
1.3 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 |
0.8 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.2 |
0.8 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
10.6 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.2 |
0.2 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.2 |
1.7 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
0.1 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.4 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.4 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.6 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
1.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.7 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
2.2 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 |
0.3 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.1 |
1.3 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
1.4 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.7 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 |
0.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.7 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.1 |
0.8 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
0.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.5 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.6 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
1.0 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.3 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
1.0 |
GO:1904754 |
positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.1 |
2.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
0.7 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.1 |
1.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.8 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.1 |
0.8 |
GO:0032196 |
transposition(GO:0032196) |
| 0.1 |
0.4 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.1 |
3.0 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.1 |
0.3 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
1.0 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.1 |
0.5 |
GO:0071105 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
1.1 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 |
0.3 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
1.0 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.1 |
0.8 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
7.4 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.4 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.1 |
1.2 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 |
2.6 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.1 |
0.5 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.9 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.7 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.3 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
0.8 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.7 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.3 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.7 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.8 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.5 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.1 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.1 |
0.6 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.1 |
GO:0010873 |
positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 |
1.6 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
1.0 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.7 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
1.3 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.4 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 |
0.7 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
1.1 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.1 |
0.4 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.1 |
0.3 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
2.6 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.1 |
1.8 |
GO:0050868 |
negative regulation of T cell activation(GO:0050868) |
| 0.1 |
0.9 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 |
0.2 |
GO:0097066 |
response to thyroid hormone(GO:0097066) |
| 0.1 |
0.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.7 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
1.0 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.3 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
1.6 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.4 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.4 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.6 |
GO:0060371 |
regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.1 |
0.4 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.3 |
GO:0090069 |
regulation of ribosome biogenesis(GO:0090069) |
| 0.1 |
1.6 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.5 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 |
0.9 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 |
0.8 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.3 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
3.4 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.1 |
0.5 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
0.5 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.1 |
0.9 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.3 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
3.1 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
0.5 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.6 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.2 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.6 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.1 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.3 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.3 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
1.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
1.6 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.4 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.5 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
0.3 |
GO:0002329 |
pre-B cell differentiation(GO:0002329) |
| 0.1 |
0.6 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 |
0.3 |
GO:0060215 |
histone H3-K4 acetylation(GO:0043973) primitive hemopoiesis(GO:0060215) primitive erythrocyte differentiation(GO:0060319) |
| 0.1 |
0.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.5 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 |
0.3 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
4.0 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.1 |
0.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.1 |
0.6 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.4 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.2 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) |
| 0.1 |
0.1 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.1 |
1.4 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
1.1 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.1 |
0.1 |
GO:0003157 |
endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) |
| 0.1 |
0.3 |
GO:0007350 |
blastoderm segmentation(GO:0007350) |
| 0.1 |
0.1 |
GO:0097680 |
double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 |
0.3 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
0.1 |
GO:0060558 |
regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.1 |
0.2 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
1.0 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.1 |
0.1 |
GO:0016056 |
response to light intensity(GO:0009642) rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.3 |
GO:1904667 |
negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 |
0.4 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.3 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.3 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.9 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.9 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.0 |
1.0 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.6 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
1.2 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.2 |
GO:0009996 |
negative regulation of cell fate specification(GO:0009996) |
| 0.0 |
0.5 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.9 |
GO:0042088 |
T-helper 1 type immune response(GO:0042088) |
| 0.0 |
1.0 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
4.2 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.2 |
GO:2000302 |
positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 |
4.0 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.5 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
1.1 |
GO:0014037 |
Schwann cell differentiation(GO:0014037) |
| 0.0 |
0.3 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 |
0.5 |
GO:0034377 |
plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 |
1.0 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 |
0.1 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.3 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.0 |
0.7 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.5 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.7 |
GO:0051351 |
positive regulation of ligase activity(GO:0051351) |
| 0.0 |
2.0 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.5 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.3 |
GO:0033151 |
V(D)J recombination(GO:0033151) |
| 0.0 |
0.6 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.0 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.0 |
0.6 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.8 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.6 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.3 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.1 |
GO:0070162 |
adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 |
0.1 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.4 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.9 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.6 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.3 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
1.1 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.4 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.3 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.3 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.2 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.3 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 |
2.3 |
GO:0031570 |
DNA integrity checkpoint(GO:0031570) |
| 0.0 |
0.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.3 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.2 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 |
1.7 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
0.3 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.2 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.4 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.2 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.1 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.0 |
0.1 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.2 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.1 |
GO:0045930 |
negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 |
0.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.6 |
GO:0044728 |
DNA methylation or demethylation(GO:0044728) |
| 0.0 |
0.5 |
GO:0031960 |
response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 |
0.1 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.9 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
1.1 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.5 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.4 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.3 |
GO:0010883 |
regulation of lipid storage(GO:0010883) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.0 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.2 |
GO:0001895 |
retina homeostasis(GO:0001895) |
| 0.0 |
0.1 |
GO:0090160 |
Golgi to lysosome transport(GO:0090160) |
| 0.0 |
0.5 |
GO:0050821 |
protein stabilization(GO:0050821) |
| 0.0 |
0.5 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
1.3 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.8 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.2 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.0 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.1 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0006924 |
activation-induced cell death of T cells(GO:0006924) |
| 0.0 |
0.3 |
GO:0090003 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 |
1.2 |
GO:0010466 |
negative regulation of peptidase activity(GO:0010466) |
| 0.0 |
0.3 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.0 |
0.5 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0048636 |
positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 |
0.4 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |