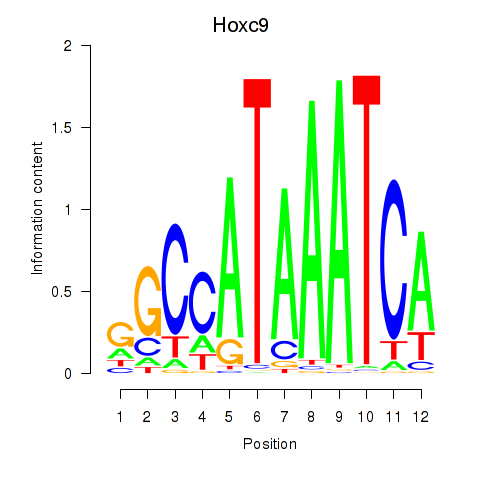
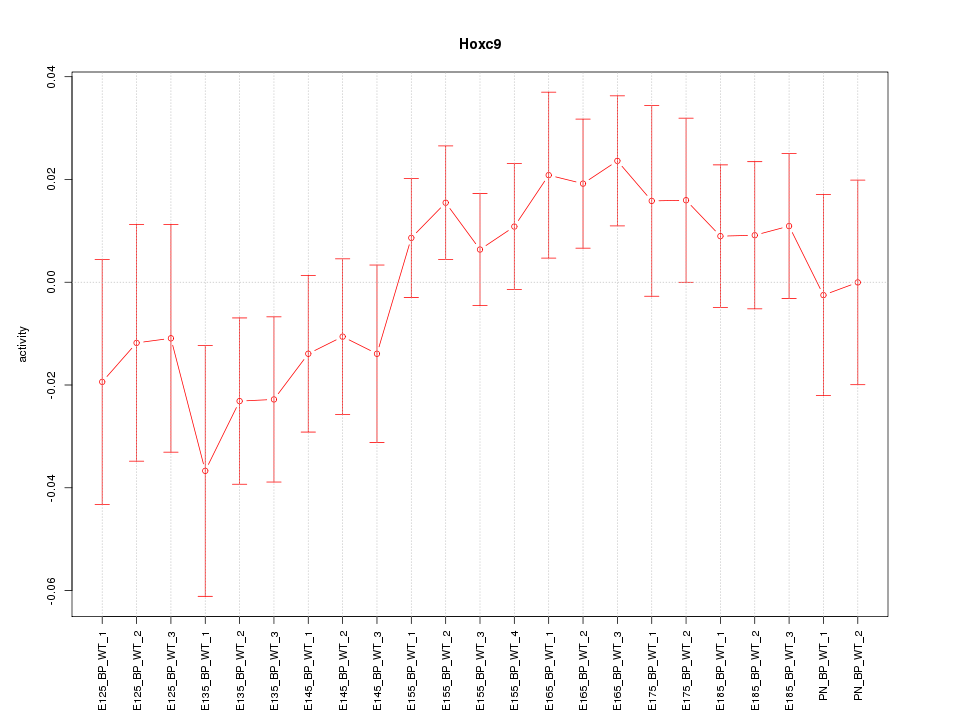
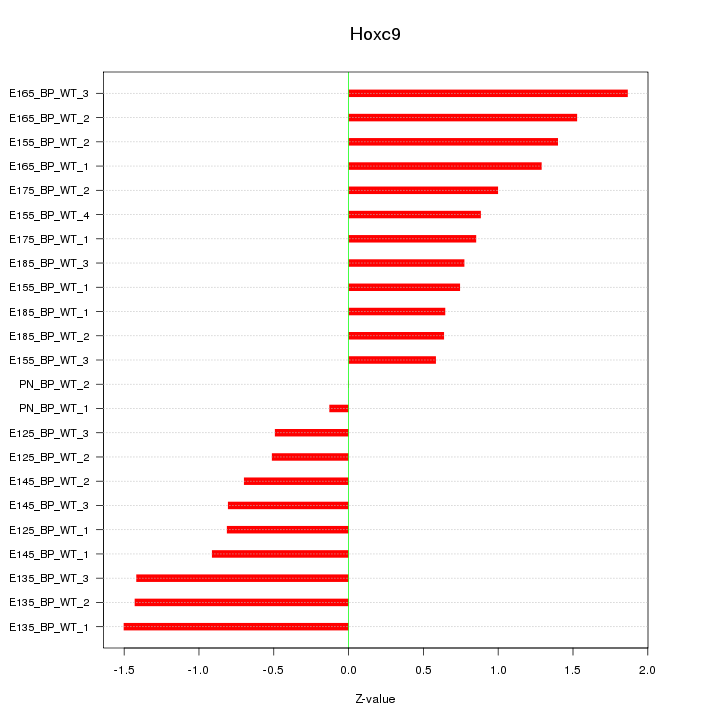
| 1.5 |
4.4 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.9 |
2.8 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 |
2.4 |
GO:0003219 |
atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 0.7 |
3.3 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.5 |
1.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 |
1.8 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.4 |
4.5 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.3 |
3.0 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.2 |
1.6 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
7.0 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.1 |
3.5 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.1 |
1.6 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
4.6 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 |
0.4 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 |
1.9 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.3 |
GO:1902524 |
positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
3.2 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.6 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
2.3 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
1.0 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) |
| 0.1 |
0.3 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 |
0.3 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.7 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.5 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.6 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
1.6 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.2 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.5 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.5 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.1 |
GO:2000768 |
glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 |
0.5 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.4 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |