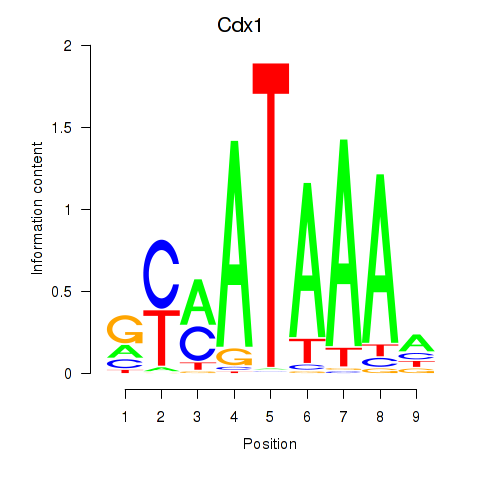
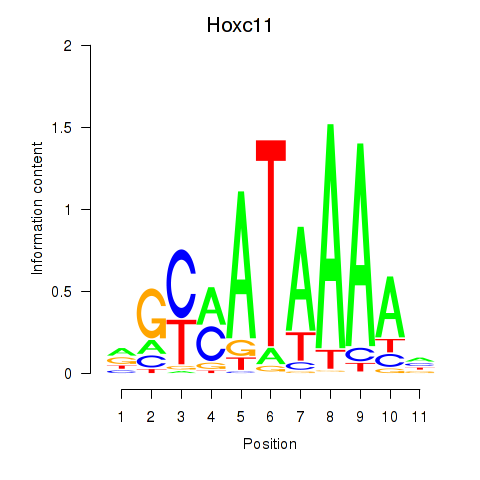
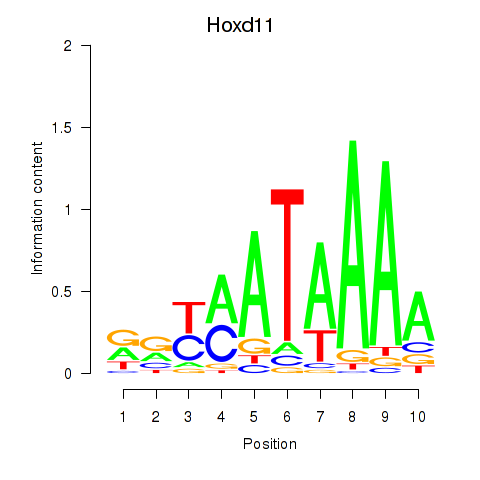
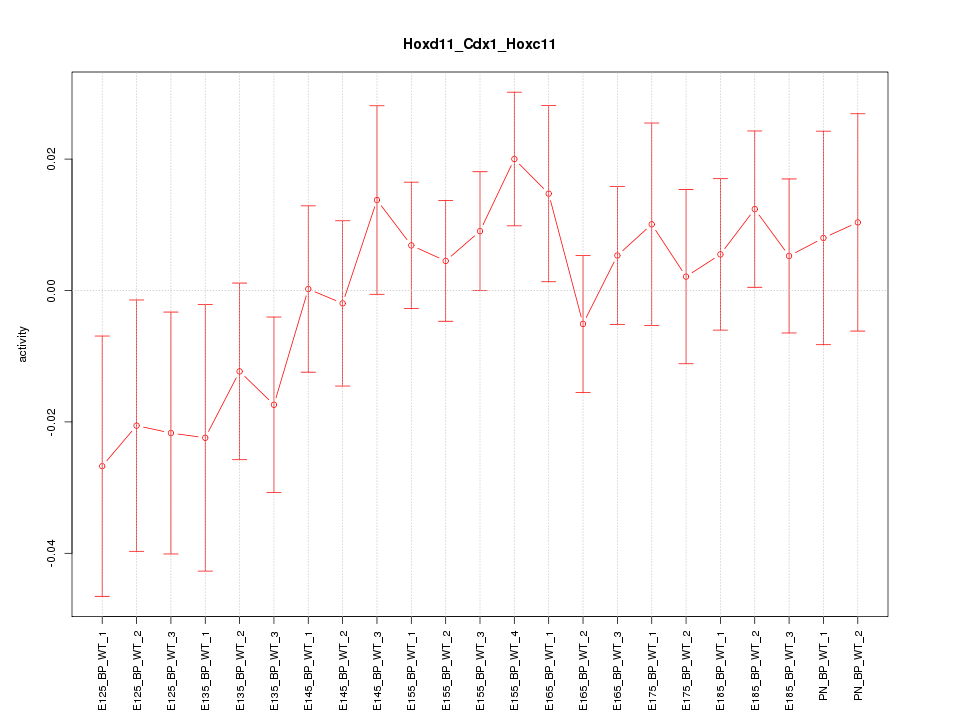
| 1.8 |
20.2 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 1.1 |
3.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.0 |
6.8 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.6 |
1.9 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 |
1.4 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 |
3.2 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.3 |
1.0 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 |
0.9 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.3 |
0.9 |
GO:1900020 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 |
4.8 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 |
0.9 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 |
2.4 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 |
5.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 |
0.5 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.3 |
1.8 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 |
1.7 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.2 |
0.9 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 |
0.7 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.2 |
0.7 |
GO:2001137 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.2 |
3.6 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.2 |
0.6 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
2.1 |
GO:1901629 |
regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 |
4.7 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
1.6 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
0.4 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.6 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.1 |
0.6 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
0.4 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.1 |
1.4 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.8 |
GO:0032196 |
transposition(GO:0032196) |
| 0.1 |
1.2 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
2.0 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.7 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.7 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
1.5 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
1.4 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.1 |
0.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
2.4 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.7 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.6 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.4 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.6 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.2 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.1 |
0.2 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 |
0.8 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.1 |
0.7 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.1 |
2.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.7 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.2 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
3.3 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.1 |
0.9 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.4 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.2 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 |
0.6 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0060729 |
pancreatic A cell differentiation(GO:0003310) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.4 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.0 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.3 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 |
0.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.3 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.1 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.4 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
1.2 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
3.7 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:1902373 |
negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 |
0.2 |
GO:0031272 |
regulation of pseudopodium assembly(GO:0031272) |
| 0.0 |
0.7 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.6 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.4 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
0.5 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0000052 |
citrulline metabolic process(GO:0000052) |
| 0.0 |
0.6 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.5 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.5 |
GO:0045471 |
response to ethanol(GO:0045471) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.8 |
GO:0030316 |
osteoclast differentiation(GO:0030316) |
| 0.0 |
2.5 |
GO:0031589 |
cell-substrate adhesion(GO:0031589) |