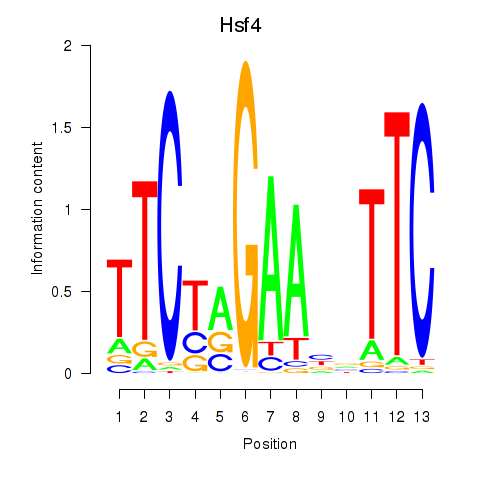
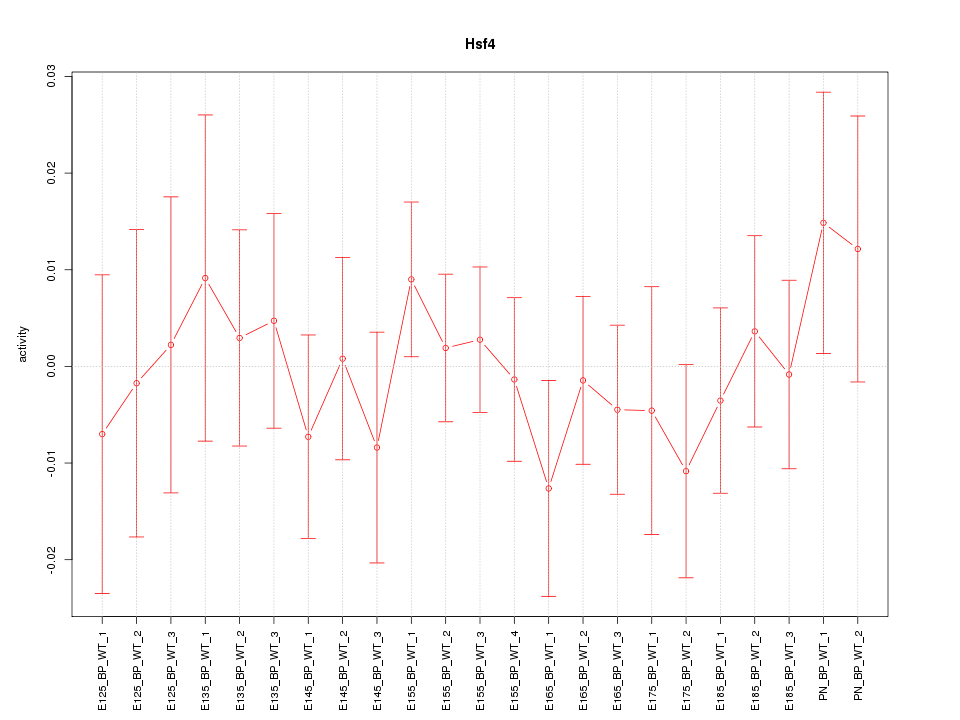
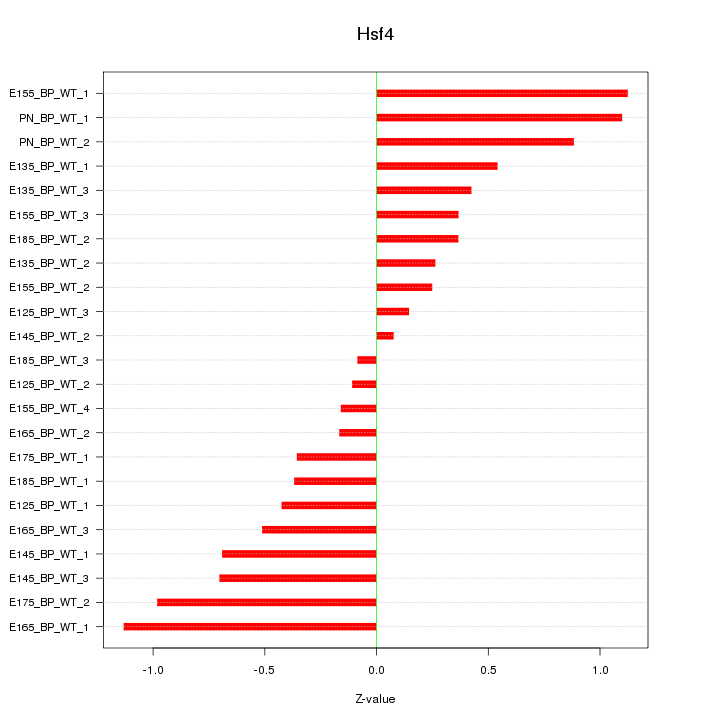
| 0.6 |
1.7 |
GO:0035799 |
ureter maturation(GO:0035799) |
| 0.4 |
1.1 |
GO:1900041 |
negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 |
1.5 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 |
0.8 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.2 |
1.3 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 |
0.6 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 |
0.5 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 |
0.5 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 |
0.9 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.4 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.4 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.1 |
0.5 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.1 |
0.3 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.1 |
0.4 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.3 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
0.5 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.4 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 |
0.3 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.1 |
0.3 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.1 |
0.2 |
GO:0035771 |
interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 |
1.0 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.3 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 |
0.6 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.2 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
0.3 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 |
0.3 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
0.1 |
GO:0060149 |
negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 |
0.2 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.4 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
0.4 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.3 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 |
0.3 |
GO:0042228 |
interleukin-8 biosynthetic process(GO:0042228) |
| 0.0 |
0.3 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.0 |
0.1 |
GO:1904154 |
protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.3 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.3 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.2 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.0 |
0.0 |
GO:0048550 |
negative regulation of pinocytosis(GO:0048550) |
| 0.0 |
0.5 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.4 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.1 |
GO:0010248 |
B cell negative selection(GO:0002352) response to mycotoxin(GO:0010046) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 |
0.2 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.3 |
GO:0051036 |
regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 |
0.3 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.1 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 |
0.8 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.3 |
GO:0090037 |
positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 |
0.1 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 |
0.2 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 |
0.1 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.0 |
0.4 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.1 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.0 |
0.1 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.1 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 |
0.2 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.4 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.3 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 |
1.0 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.1 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.1 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.0 |
0.2 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.3 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 |
0.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
0.2 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.2 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.2 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.3 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.0 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.0 |
0.1 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.1 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.0 |
0.5 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.2 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.2 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.1 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |