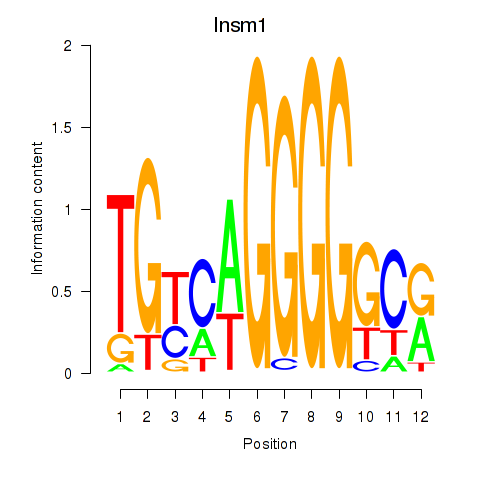
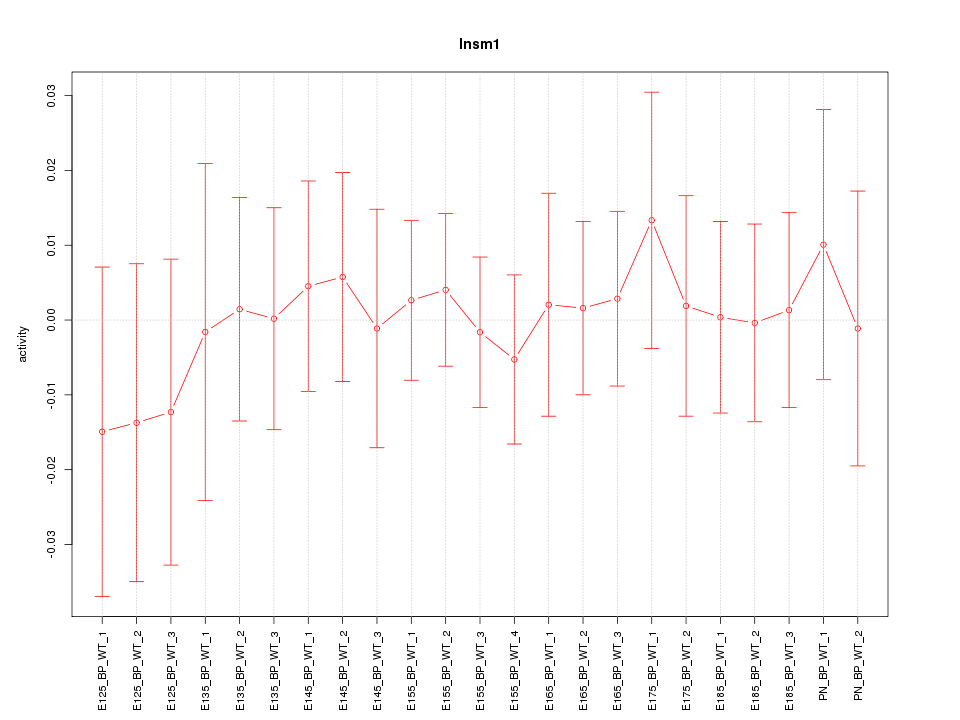
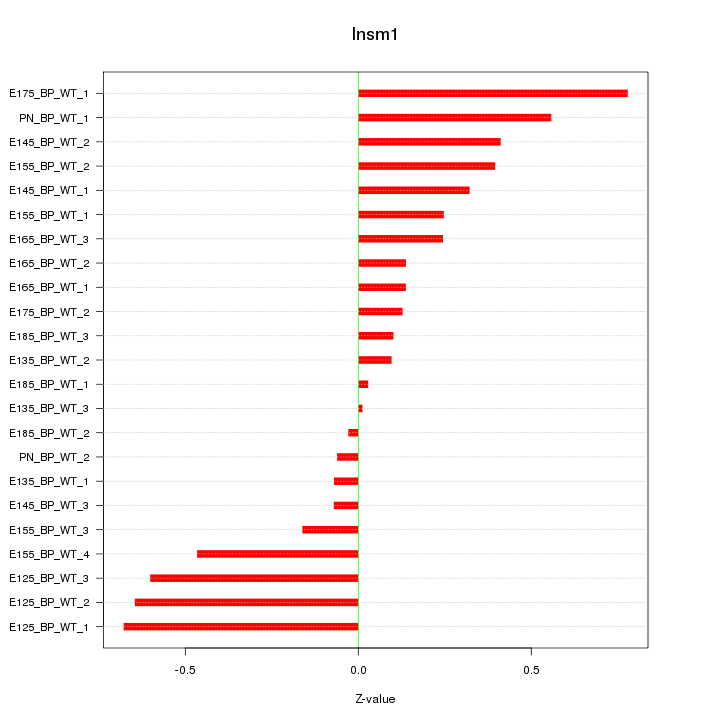
| 0.4 |
1.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.3 |
0.9 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 |
0.6 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.9 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.6 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.4 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.5 |
GO:1905206 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 |
0.4 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.3 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.6 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.5 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.2 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
0.2 |
GO:0061428 |
positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.2 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 |
0.2 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 |
0.4 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.1 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 |
0.2 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 |
0.3 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
1.0 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.5 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.0 |
1.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.0 |
0.1 |
GO:0089700 |
positive regulation of T cell receptor signaling pathway(GO:0050862) protein kinase D signaling(GO:0089700) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.4 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.1 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.0 |
0.1 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.4 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.0 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 |
0.0 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
0.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.0 |
0.4 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.2 |
GO:0030203 |
glycosaminoglycan metabolic process(GO:0030203) |