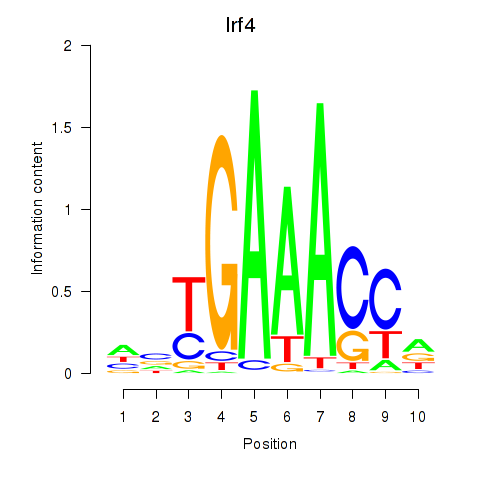
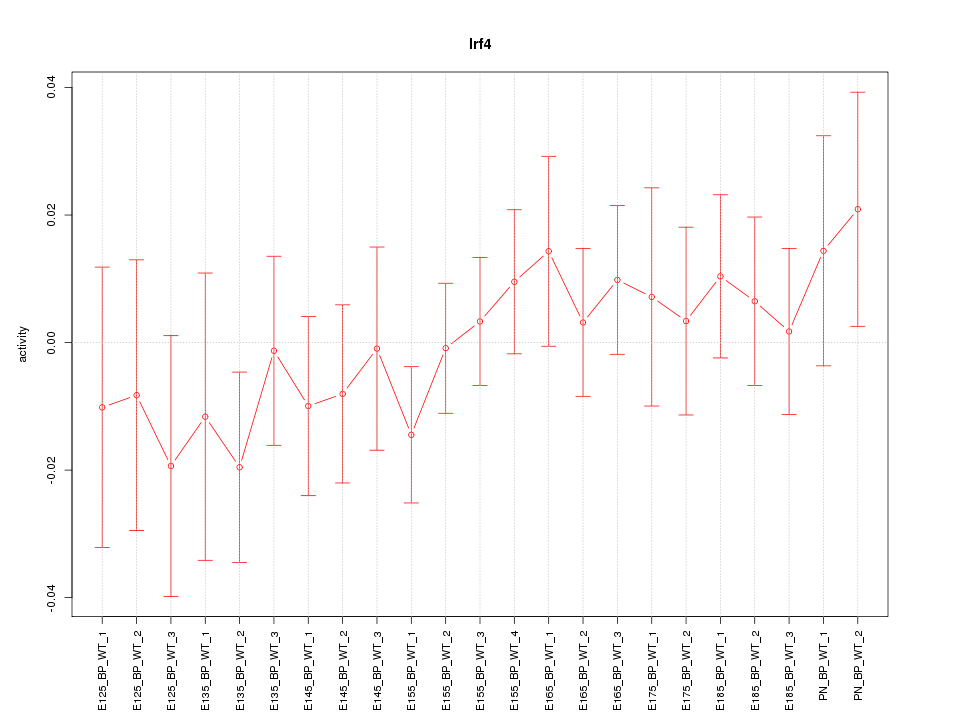
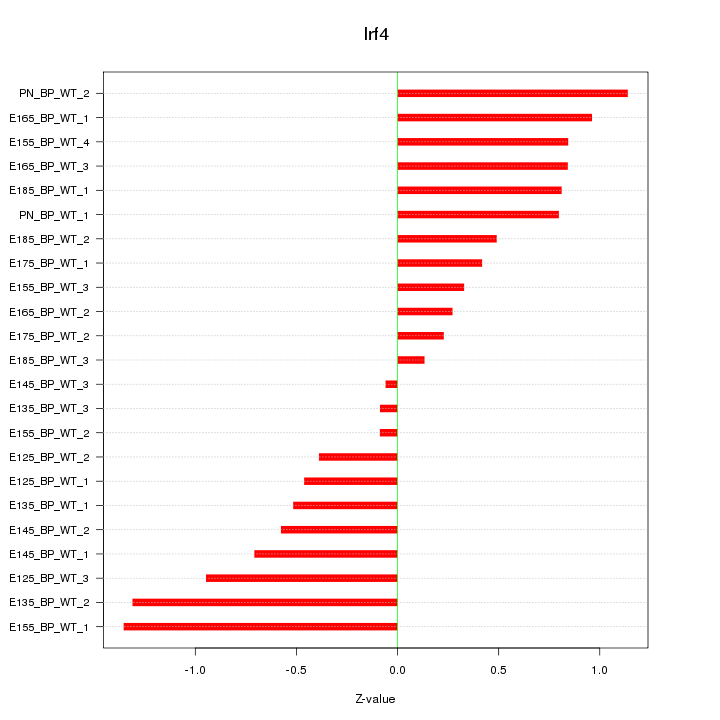
| 1.1 |
3.2 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.7 |
2.1 |
GO:1900041 |
intestinal D-glucose absorption(GO:0001951) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.5 |
1.6 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 |
1.8 |
GO:0003430 |
tolerance induction to self antigen(GO:0002513) growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.4 |
1.9 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 |
1.5 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 |
1.5 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.3 |
2.9 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.3 |
1.7 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 |
0.9 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.2 |
1.8 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.2 |
0.5 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
1.9 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
1.2 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
1.8 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 |
0.9 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.8 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.1 |
0.6 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.5 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
1.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.6 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.5 |
GO:0030832 |
regulation of actin filament length(GO:0030832) |
| 0.1 |
0.6 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
1.0 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.3 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.8 |
GO:0032695 |
negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 |
0.2 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.3 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.2 |
GO:0035799 |
ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 |
0.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.9 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.8 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.3 |
GO:0000429 |
carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 |
4.4 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
1.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 |
0.4 |
GO:0031670 |
cellular response to nutrient(GO:0031670) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.3 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.1 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 |
0.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.5 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.1 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 |
0.7 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.1 |
GO:0032621 |
interleukin-18 production(GO:0032621) positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 |
0.8 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.0 |
GO:2000620 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.2 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |