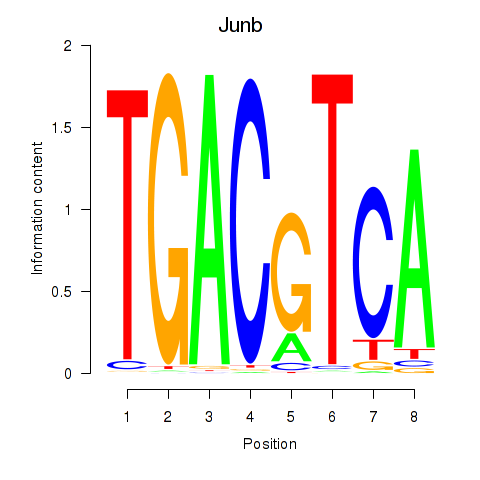
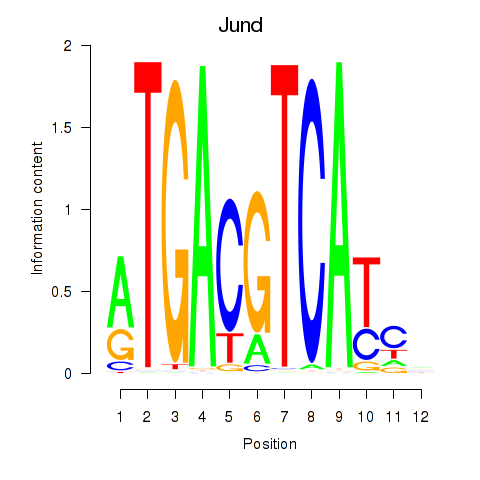
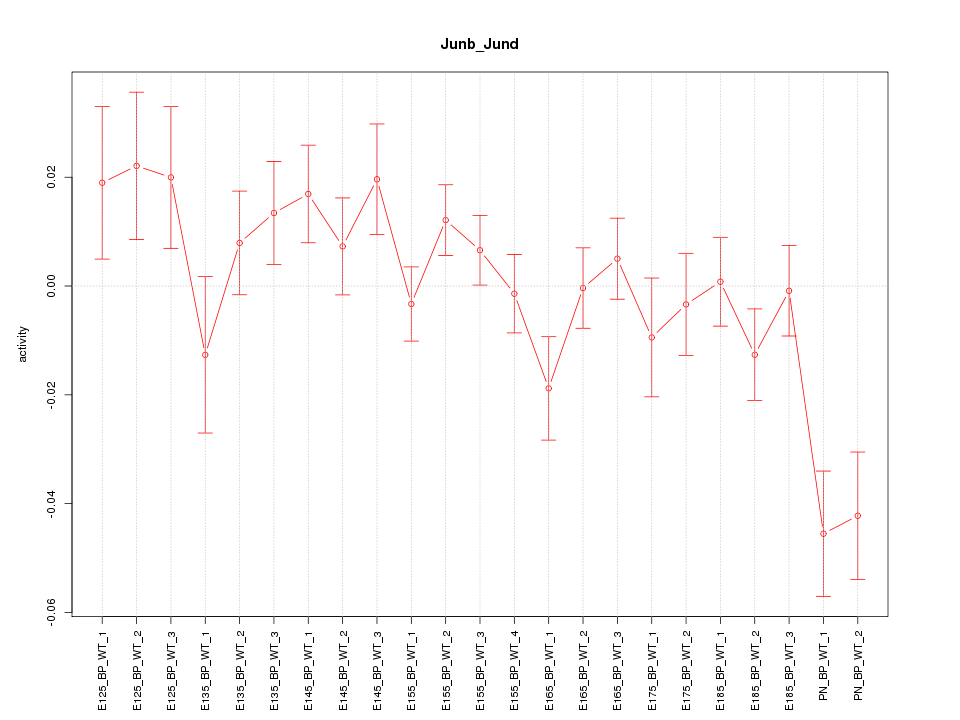
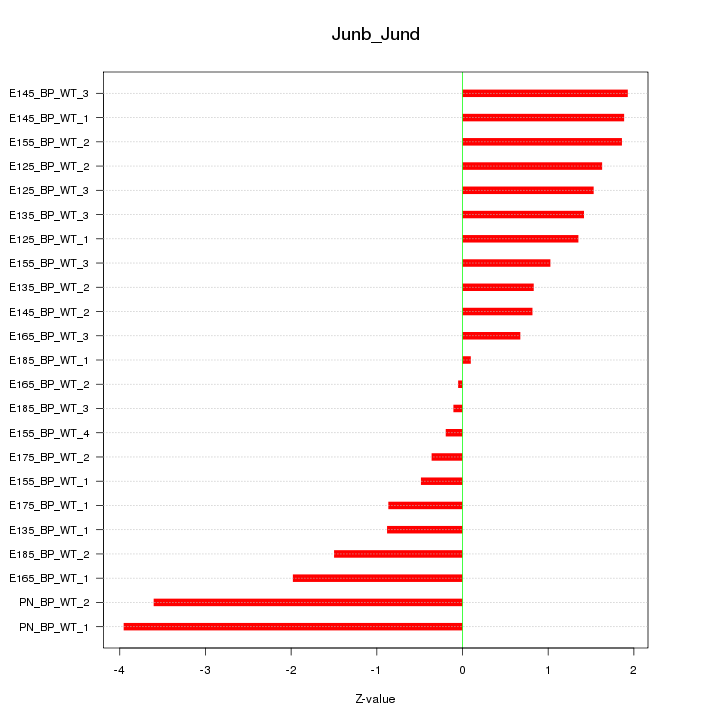
| 3.7 |
11.0 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 2.9 |
17.3 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.3 |
6.8 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.6 |
4.8 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 1.3 |
3.8 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 1.2 |
5.0 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.1 |
3.3 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 1.1 |
3.3 |
GO:0030070 |
insulin processing(GO:0030070) |
| 1.0 |
2.9 |
GO:0060067 |
cervix development(GO:0060067) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.9 |
2.8 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.9 |
2.7 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.9 |
2.6 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.9 |
3.4 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.8 |
3.4 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.8 |
3.0 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.8 |
2.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.7 |
3.7 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.7 |
2.1 |
GO:0071649 |
regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.7 |
2.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.7 |
0.7 |
GO:2001180 |
negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.6 |
1.9 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.6 |
1.8 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.6 |
4.1 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.6 |
10.2 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.6 |
1.7 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.5 |
1.6 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.5 |
4.9 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 |
4.8 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 |
2.7 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.5 |
1.4 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 |
7.2 |
GO:0061162 |
establishment of monopolar cell polarity(GO:0061162) |
| 0.5 |
1.9 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.5 |
1.4 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.4 |
1.3 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.4 |
1.3 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 |
1.3 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.4 |
3.6 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 |
4.4 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 |
4.3 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.4 |
3.9 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.4 |
1.1 |
GO:0060364 |
frontal suture morphogenesis(GO:0060364) |
| 0.4 |
0.4 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.4 |
3.6 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 |
3.2 |
GO:0086018 |
SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.4 |
2.8 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.4 |
2.8 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.3 |
2.4 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.3 |
1.0 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.3 |
2.7 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 |
1.4 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.3 |
8.1 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.3 |
2.8 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.3 |
0.9 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.3 |
3.1 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 |
0.8 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 |
5.0 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.3 |
3.8 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.3 |
1.1 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.3 |
2.9 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.3 |
1.3 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.3 |
0.8 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 |
2.0 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.3 |
0.8 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 |
3.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.2 |
1.9 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.2 |
0.2 |
GO:0044262 |
cellular carbohydrate metabolic process(GO:0044262) |
| 0.2 |
0.9 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 |
1.6 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.2 |
0.9 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 |
1.6 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 |
0.9 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.2 |
0.9 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.2 |
0.6 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.2 |
1.5 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 |
0.8 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 |
0.8 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 |
0.8 |
GO:0060459 |
extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 |
0.6 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.2 |
0.8 |
GO:0046618 |
drug export(GO:0046618) |
| 0.2 |
2.0 |
GO:0006706 |
steroid catabolic process(GO:0006706) negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 |
1.2 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.2 |
0.8 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.2 |
0.4 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.2 |
0.6 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 |
6.5 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.2 |
0.6 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 |
0.6 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.2 |
0.6 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
1.3 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 |
0.4 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.2 |
1.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.2 |
0.7 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.2 |
1.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 |
0.9 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.2 |
0.5 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.2 |
0.8 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 |
1.5 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 |
0.6 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 |
4.8 |
GO:0034067 |
protein localization to Golgi apparatus(GO:0034067) |
| 0.2 |
0.8 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.2 |
0.5 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.1 |
0.7 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.7 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
0.7 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
0.3 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.1 |
2.0 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
1.4 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.4 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.7 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 |
1.0 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.4 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.1 |
1.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 |
0.7 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.1 |
0.4 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
1.2 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.1 |
0.5 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
0.8 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.5 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 |
0.8 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.1 |
0.5 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
2.1 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.1 |
0.4 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.5 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 |
2.9 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.6 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.1 |
0.5 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 |
0.5 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 |
0.3 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
1.1 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 |
0.6 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
0.5 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 |
1.8 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
0.3 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 |
0.1 |
GO:0007135 |
meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 |
1.5 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 |
0.4 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.8 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) |
| 0.1 |
0.3 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
3.0 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 |
2.0 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.5 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 |
3.0 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.1 |
0.4 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
0.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.5 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.5 |
GO:0032230 |
positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.1 |
0.6 |
GO:0070365 |
hepatocyte differentiation(GO:0070365) |
| 0.1 |
1.3 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.4 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 |
0.6 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 |
0.5 |
GO:0034115 |
negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.2 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 |
0.9 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 |
1.3 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.1 |
1.9 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.8 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 |
1.0 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.4 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
1.4 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 |
0.3 |
GO:0032025 |
response to cobalt ion(GO:0032025) |
| 0.1 |
1.6 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 |
1.0 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.1 |
1.3 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 |
0.3 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 |
0.3 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 |
1.2 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.3 |
GO:1901723 |
negative regulation of cell proliferation involved in kidney development(GO:1901723) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 |
0.9 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.1 |
0.2 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 |
0.6 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
0.8 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
1.1 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 |
0.6 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.7 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.1 |
0.5 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 |
1.0 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
1.7 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
0.8 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 |
0.8 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.1 |
0.3 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
1.4 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
0.4 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.4 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.6 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.1 |
1.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.1 |
0.5 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
1.4 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
25.8 |
GO:0043161 |
proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.1 |
0.6 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.1 |
9.4 |
GO:0010466 |
negative regulation of peptidase activity(GO:0010466) |
| 0.1 |
0.4 |
GO:0003418 |
growth plate cartilage chondrocyte differentiation(GO:0003418) growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 |
2.9 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.1 |
0.3 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.4 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
0.9 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
2.0 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.1 |
0.3 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.1 |
0.7 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 |
1.3 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.1 |
0.5 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.3 |
GO:0032811 |
regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 |
0.6 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.1 |
1.6 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.1 |
0.8 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.1 |
0.6 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.3 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.1 |
3.8 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.3 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.1 |
2.0 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.4 |
GO:0090051 |
regulation of skeletal muscle fiber development(GO:0048742) negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.3 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.5 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.8 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.4 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.9 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.1 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.1 |
GO:0010886 |
regulation of cholesterol storage(GO:0010885) positive regulation of cholesterol storage(GO:0010886) |
| 0.0 |
0.2 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) regulation of lipoprotein lipase activity(GO:0051004) negative regulation of lipase activity(GO:0060192) |
| 0.0 |
0.1 |
GO:0072395 |
signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 |
0.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.3 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.6 |
GO:0003094 |
glomerular filtration(GO:0003094) |
| 0.0 |
1.5 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.7 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
0.6 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
1.1 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 |
1.2 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.3 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.0 |
0.3 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.3 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.2 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.6 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.3 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.3 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.0 |
1.7 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
1.3 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.9 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
1.8 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.3 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.4 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.9 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.5 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
1.0 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
2.5 |
GO:0007612 |
learning(GO:0007612) |
| 0.0 |
0.2 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
1.7 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
1.6 |
GO:0007215 |
glutamate receptor signaling pathway(GO:0007215) |
| 0.0 |
2.2 |
GO:0031110 |
regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 |
0.7 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.0 |
0.4 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.1 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.1 |
GO:2000189 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.6 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 |
0.9 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.4 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.2 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.9 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0021592 |
fourth ventricle development(GO:0021592) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.5 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.0 |
0.2 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.1 |
GO:0016560 |
fatty acid alpha-oxidation(GO:0001561) protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 |
0.2 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.4 |
GO:0006584 |
catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 |
0.9 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.2 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.0 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.6 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.4 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
1.0 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 |
0.1 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.5 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.4 |
GO:0043242 |
negative regulation of protein complex disassembly(GO:0043242) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 |
0.2 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.1 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 |
0.6 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.4 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.1 |
GO:0010801 |
negative regulation of peptidyl-threonine phosphorylation(GO:0010801) positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.7 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.0 |
GO:0070099 |
regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |