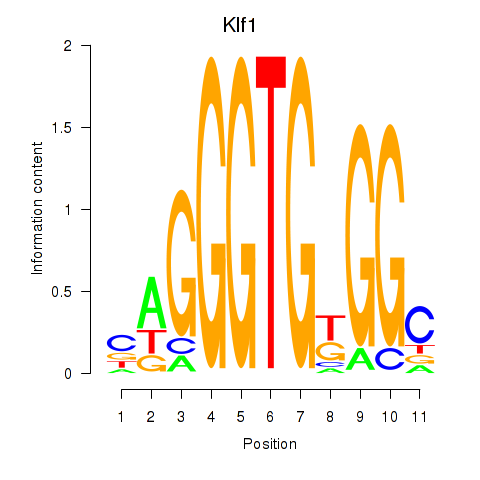
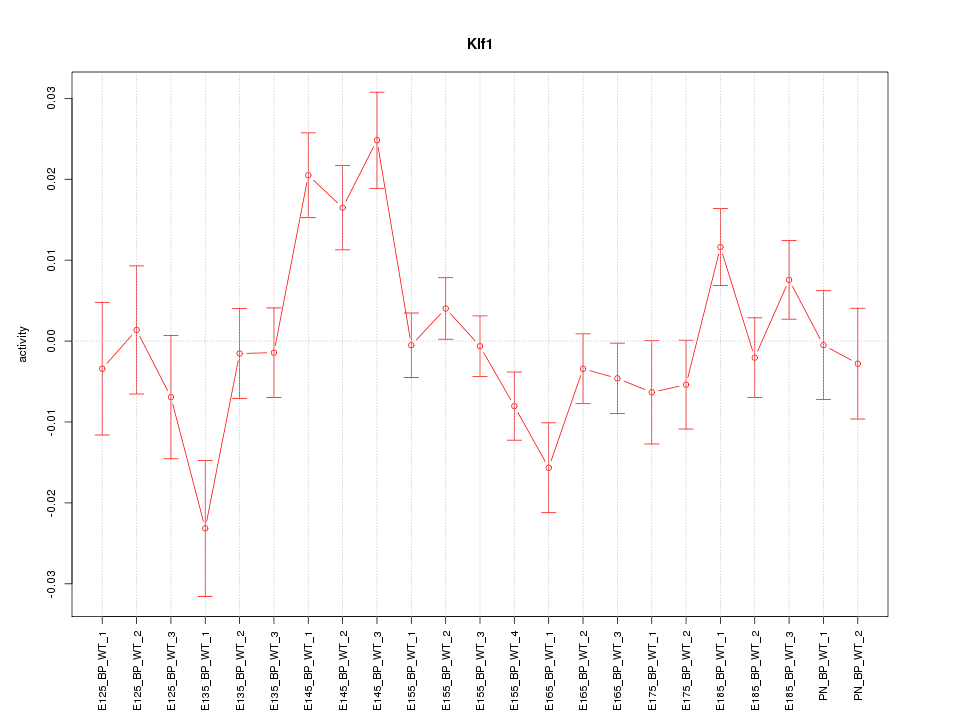
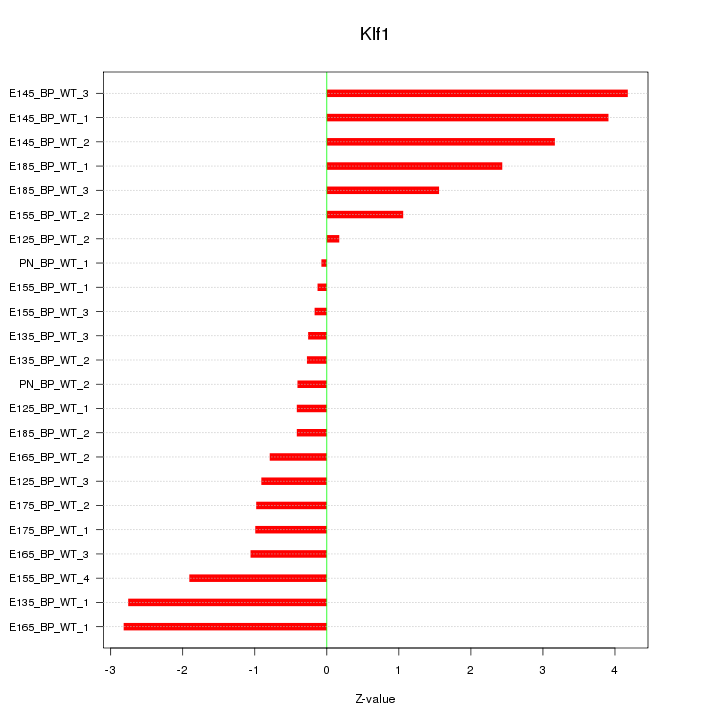
| 1.2 |
3.6 |
GO:0046959 |
habituation(GO:0046959) |
| 1.1 |
3.4 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.8 |
5.8 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.8 |
2.4 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.8 |
2.3 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.7 |
4.0 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.6 |
1.9 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.6 |
3.0 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 |
2.2 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.5 |
3.2 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.5 |
1.5 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.5 |
1.5 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.5 |
1.5 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 |
2.8 |
GO:0014054 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.5 |
1.4 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 |
2.1 |
GO:0042321 |
negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) |
| 0.4 |
4.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.4 |
2.4 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 |
1.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 |
1.8 |
GO:0021636 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.4 |
5.7 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 |
0.3 |
GO:0071280 |
cellular response to copper ion(GO:0071280) |
| 0.3 |
4.8 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 |
0.3 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.3 |
1.3 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.3 |
1.0 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 |
1.0 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.3 |
1.0 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 |
1.3 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.3 |
3.4 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 |
1.5 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
1.2 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.3 |
0.6 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 |
1.2 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.3 |
1.5 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 |
0.3 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.3 |
2.9 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 |
0.9 |
GO:2000097 |
regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 |
0.3 |
GO:0044851 |
hair cycle phase(GO:0044851) hair follicle maturation(GO:0048820) |
| 0.3 |
1.5 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.3 |
0.9 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.3 |
1.7 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
3.9 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.3 |
0.8 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 |
0.8 |
GO:0097401 |
synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 |
0.8 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 |
0.3 |
GO:0032902 |
nerve growth factor production(GO:0032902) |
| 0.3 |
0.8 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.3 |
0.5 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.3 |
2.0 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 |
0.5 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 |
1.0 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 |
0.7 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.2 |
1.4 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.2 |
0.7 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.5 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.2 |
1.6 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.2 |
0.7 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 |
0.6 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.2 |
0.9 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.2 |
1.5 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
1.5 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 |
1.6 |
GO:0021894 |
cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.2 |
2.0 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.2 |
2.8 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.2 |
0.6 |
GO:0032233 |
positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 |
1.0 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 |
0.2 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.2 |
0.8 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
0.8 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.2 |
1.8 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 |
0.8 |
GO:0045794 |
negative regulation of cell volume(GO:0045794) |
| 0.2 |
0.6 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 |
1.7 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.2 |
0.7 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 |
0.5 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.2 |
0.5 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.2 |
0.5 |
GO:0046878 |
negative regulation of urine volume(GO:0035811) positive regulation of saliva secretion(GO:0046878) |
| 0.2 |
0.5 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 |
1.6 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.2 |
0.5 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 |
0.7 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.2 |
1.9 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.2 |
0.3 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
0.5 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 |
0.5 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 |
1.0 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 |
0.5 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.2 |
0.5 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 |
0.5 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 |
0.3 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.2 |
0.5 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 |
0.6 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.2 |
0.6 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.2 |
1.4 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.2 |
0.5 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 |
0.6 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.2 |
0.6 |
GO:0014045 |
establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.2 |
0.6 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
0.8 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.2 |
0.5 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 |
0.5 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 |
0.6 |
GO:0046709 |
IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.2 |
0.9 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.2 |
0.5 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.6 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.7 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.6 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 |
1.0 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.1 |
0.3 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 |
0.9 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.6 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.9 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 |
1.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.1 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 |
0.4 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.1 |
0.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.7 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.1 |
0.6 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 |
1.9 |
GO:0006677 |
glycosylceramide metabolic process(GO:0006677) |
| 0.1 |
0.8 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.7 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
2.6 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
1.0 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 |
1.0 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.1 |
0.4 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
0.4 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 |
0.5 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 |
0.7 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.3 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.7 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.9 |
GO:0034969 |
histone arginine methylation(GO:0034969) |
| 0.1 |
2.0 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
3.2 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
2.1 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.1 |
0.7 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.1 |
GO:0061738 |
late endosomal microautophagy(GO:0061738) |
| 0.1 |
0.5 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 |
0.4 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.9 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.4 |
GO:2001170 |
positive regulation of fat cell proliferation(GO:0070346) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
0.4 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 |
0.5 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.1 |
0.8 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.6 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.4 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.1 |
0.4 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.5 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.2 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.1 |
1.7 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 |
0.4 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.4 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.1 |
0.5 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.1 |
0.2 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.4 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.1 |
0.7 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 |
0.9 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
0.2 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.5 |
GO:0061428 |
positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
0.3 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.1 |
0.2 |
GO:1903243 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 |
0.5 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
3.4 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.1 |
0.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.6 |
GO:1902856 |
negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
1.6 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.5 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.2 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.1 |
0.5 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
0.9 |
GO:0021681 |
cerebellar granular layer development(GO:0021681) |
| 0.1 |
3.0 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.1 |
0.8 |
GO:0014832 |
urinary bladder smooth muscle contraction(GO:0014832) |
| 0.1 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
2.1 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.1 |
GO:0051695 |
actin filament uncapping(GO:0051695) |
| 0.1 |
0.7 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.1 |
0.4 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.1 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.3 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 |
1.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.4 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.2 |
GO:0032536 |
intestinal D-glucose absorption(GO:0001951) regulation of microvillus length(GO:0032532) regulation of cell projection size(GO:0032536) terminal web assembly(GO:1902896) |
| 0.1 |
0.5 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 |
0.2 |
GO:0030969 |
mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.1 |
0.4 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 |
1.5 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.1 |
0.5 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.1 |
0.4 |
GO:0051006 |
positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.1 |
0.5 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.6 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
0.3 |
GO:1903984 |
regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 |
2.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.4 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
0.3 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 |
0.5 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 |
0.3 |
GO:0070602 |
regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 |
0.2 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.6 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.1 |
1.3 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.3 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
2.0 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.1 |
0.4 |
GO:0090035 |
regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 |
0.6 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.5 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
0.8 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.8 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.1 |
0.4 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.6 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
0.3 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.1 |
1.3 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.5 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
0.4 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.1 |
0.3 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 |
0.5 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
2.3 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.1 |
0.3 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 |
1.1 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 |
0.2 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
0.8 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.4 |
GO:0001765 |
membrane raft assembly(GO:0001765) |
| 0.1 |
0.4 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.1 |
0.4 |
GO:0043416 |
regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 |
3.3 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
0.1 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.5 |
GO:0071472 |
cellular response to salt stress(GO:0071472) |
| 0.1 |
0.5 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
0.2 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 |
0.6 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.8 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.4 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 |
0.4 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.8 |
GO:1902170 |
cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 |
0.7 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.2 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 |
0.6 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
1.2 |
GO:0051560 |
mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 |
0.5 |
GO:0046103 |
ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 |
0.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.3 |
GO:0007227 |
signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.4 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 |
0.2 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 |
0.5 |
GO:2000786 |
positive regulation of vacuole organization(GO:0044090) positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.2 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 |
0.5 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
0.5 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.2 |
GO:1990144 |
intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.1 |
0.8 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.1 |
0.2 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 |
0.2 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 0.1 |
1.2 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.1 |
0.2 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 |
0.7 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.1 |
0.5 |
GO:0045762 |
positive regulation of adenylate cyclase activity(GO:0045762) positive regulation of lyase activity(GO:0051349) |
| 0.1 |
0.2 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 |
0.2 |
GO:0010886 |
regulation of cholesterol storage(GO:0010885) positive regulation of cholesterol storage(GO:0010886) |
| 0.1 |
0.8 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.5 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 |
0.6 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 |
0.3 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
0.6 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.2 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.1 |
0.5 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.1 |
0.1 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.1 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.4 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.4 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.2 |
GO:0046880 |
gonadotropin secretion(GO:0032274) regulation of gonadotropin secretion(GO:0032276) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.2 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 |
0.3 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
1.2 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.1 |
0.8 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.2 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 |
1.0 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.1 |
0.1 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 |
0.4 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
0.3 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 |
0.4 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.8 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.3 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.1 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 |
0.2 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 |
0.3 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) glycosphingolipid catabolic process(GO:0046479) ceramide catabolic process(GO:0046514) |
| 0.1 |
0.3 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.1 |
1.0 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 |
1.1 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.1 |
0.5 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.1 |
0.1 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.1 |
0.3 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 |
1.7 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 |
0.3 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.3 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
1.2 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.1 |
0.2 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
2.8 |
GO:0006778 |
porphyrin-containing compound metabolic process(GO:0006778) |
| 0.1 |
0.4 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.2 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.7 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.1 |
0.7 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.2 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
1.0 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.7 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.1 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.1 |
0.2 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 |
0.3 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 |
2.2 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 |
0.9 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.5 |
GO:0070207 |
protein homotrimerization(GO:0070207) |
| 0.1 |
0.1 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.1 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.1 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
0.4 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.1 |
0.5 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 |
0.2 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.2 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.7 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.1 |
0.5 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 |
0.1 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 |
0.3 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 |
0.6 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.1 |
0.5 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.2 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.8 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.1 |
1.5 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.1 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.1 |
0.3 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.1 |
1.1 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.3 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.2 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
1.0 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.6 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.7 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.3 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.1 |
0.5 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.1 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.1 |
GO:0009950 |
dorsal/ventral axis specification(GO:0009950) |
| 0.1 |
0.6 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 |
0.2 |
GO:0034124 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 |
1.5 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
0.2 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 |
0.1 |
GO:0006848 |
pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 |
0.3 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.6 |
GO:0060561 |
apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.2 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 |
0.3 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.2 |
GO:0002865 |
negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.1 |
0.2 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.1 |
0.1 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 |
0.2 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 |
0.2 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.3 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
1.0 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
0.4 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.1 |
0.4 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.6 |
GO:0031498 |
chromatin disassembly(GO:0031498) |
| 0.1 |
0.6 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.1 |
0.3 |
GO:2000650 |
negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.3 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.1 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:1904430 |
negative regulation of t-circle formation(GO:1904430) |
| 0.0 |
1.5 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.3 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.4 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.5 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 |
0.2 |
GO:0000469 |
cleavage involved in rRNA processing(GO:0000469) |
| 0.0 |
1.8 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.0 |
0.2 |
GO:2001045 |
negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.2 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
1.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.3 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.1 |
GO:0009946 |
optic vesicle morphogenesis(GO:0003404) proximal/distal axis specification(GO:0009946) neuroblast migration(GO:0097402) |
| 0.0 |
0.6 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.7 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.2 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.0 |
0.7 |
GO:0045686 |
negative regulation of glial cell differentiation(GO:0045686) |
| 0.0 |
0.2 |
GO:2000630 |
positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 |
0.1 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.2 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.1 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 |
1.0 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0021892 |
cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 |
0.2 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
1.4 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.0 |
0.4 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.2 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.9 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.0 |
0.2 |
GO:0060287 |
epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
0.2 |
GO:0021540 |
corpus callosum morphogenesis(GO:0021540) |
| 0.0 |
0.5 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 |
0.1 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.0 |
0.5 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.8 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
0.1 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.0 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.2 |
GO:0050855 |
regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.9 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.6 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.3 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
1.0 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 |
0.1 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
1.8 |
GO:0035904 |
aorta development(GO:0035904) |
| 0.0 |
0.1 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.5 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.5 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.0 |
0.1 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.7 |
GO:2000772 |
regulation of cellular senescence(GO:2000772) |
| 0.0 |
1.1 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.4 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.2 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.4 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.4 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
1.1 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:1901020 |
negative regulation of calcium ion transmembrane transporter activity(GO:1901020) |
| 0.0 |
0.1 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) |
| 0.0 |
0.2 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.1 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 |
0.5 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.4 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.2 |
GO:0045830 |
positive regulation of isotype switching(GO:0045830) |
| 0.0 |
0.1 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
0.1 |
GO:0051024 |
positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 |
0.1 |
GO:0045655 |
regulation of monocyte differentiation(GO:0045655) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.3 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.8 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0035751 |
regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 |
0.2 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.4 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.3 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.0 |
0.3 |
GO:0046036 |
CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.4 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
2.9 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.2 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
1.7 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.0 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.2 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.3 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.0 |
0.4 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.2 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
1.0 |
GO:0003254 |
regulation of membrane depolarization(GO:0003254) |
| 0.0 |
0.1 |
GO:0036258 |
multivesicular body organization(GO:0036257) multivesicular body assembly(GO:0036258) |
| 0.0 |
0.2 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 |
0.1 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.0 |
0.5 |
GO:0034067 |
protein localization to Golgi apparatus(GO:0034067) |
| 0.0 |
0.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.5 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.5 |
GO:0072384 |
organelle transport along microtubule(GO:0072384) |
| 0.0 |
0.1 |
GO:0019081 |
viral translation(GO:0019081) |
| 0.0 |
0.3 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.3 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.1 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.3 |
GO:0048753 |
pigment granule organization(GO:0048753) |
| 0.0 |
0.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.4 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.4 |
GO:0019935 |
cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 |
0.4 |
GO:0035773 |
insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.2 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.1 |
GO:0090237 |
regulation of arachidonic acid secretion(GO:0090237) |
| 0.0 |
0.1 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.5 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.4 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.1 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.2 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.0 |
0.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.0 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.4 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.5 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.2 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.4 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.1 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.5 |
GO:0002209 |
behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.1 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.1 |
GO:0001991 |
regulation of systemic arterial blood pressure by circulatory renin-angiotensin(GO:0001991) |
| 0.0 |
0.5 |
GO:0016556 |
mRNA modification(GO:0016556) |
| 0.0 |
0.2 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.3 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.7 |
GO:0007187 |
G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 |
0.3 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.0 |
GO:0072074 |
kidney mesenchyme development(GO:0072074) mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 |
0.5 |
GO:0031063 |
regulation of histone deacetylation(GO:0031063) |
| 0.0 |
0.2 |
GO:0046039 |
GTP metabolic process(GO:0046039) |
| 0.0 |
0.2 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
1.0 |
GO:0006073 |
glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 |
0.1 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.0 |
GO:0002323 |
natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 |
1.3 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.2 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.3 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
1.0 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
2.9 |
GO:0051260 |
protein homooligomerization(GO:0051260) |
| 0.0 |
0.3 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 |
0.2 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.3 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.1 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.3 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.1 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.1 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.2 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.3 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.2 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
1.6 |
GO:0002230 |
positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 |
0.0 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.0 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.2 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.2 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.0 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.1 |
GO:0009048 |
dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.3 |
GO:0071514 |
genetic imprinting(GO:0071514) |
| 0.0 |
0.0 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.3 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.1 |
GO:0052490 |
suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) modulation of programmed cell death in other organism(GO:0044531) modulation of apoptotic process in other organism(GO:0044532) response to cycloheximide(GO:0046898) modulation by symbiont of host programmed cell death(GO:0052040) negative regulation by symbiont of host programmed cell death(GO:0052041) modulation by symbiont of host apoptotic process(GO:0052150) modulation of programmed cell death in other organism involved in symbiotic interaction(GO:0052248) modulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052433) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 |
0.0 |
GO:0090656 |
t-circle formation(GO:0090656) |
| 0.0 |
0.2 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.1 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 |
0.1 |
GO:0071711 |
basement membrane organization(GO:0071711) |
| 0.0 |
0.3 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.5 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.2 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.0 |
0.1 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.3 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.0 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.4 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.0 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.2 |
GO:0015698 |
inorganic anion transport(GO:0015698) |