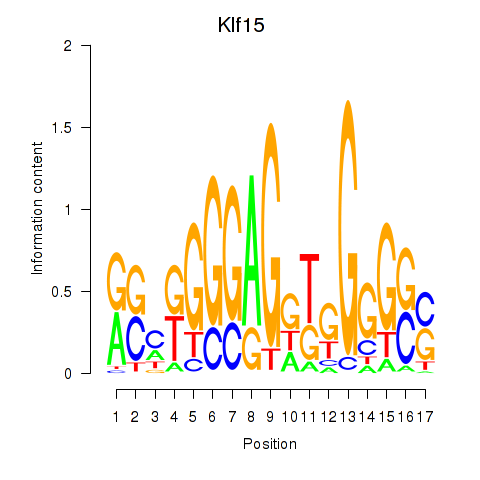
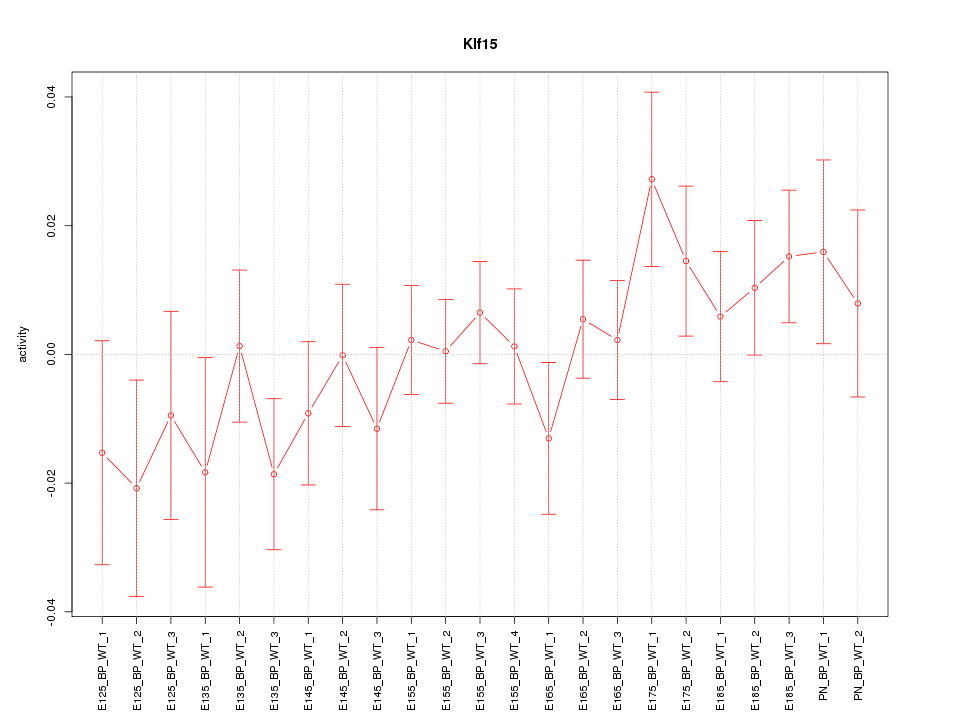
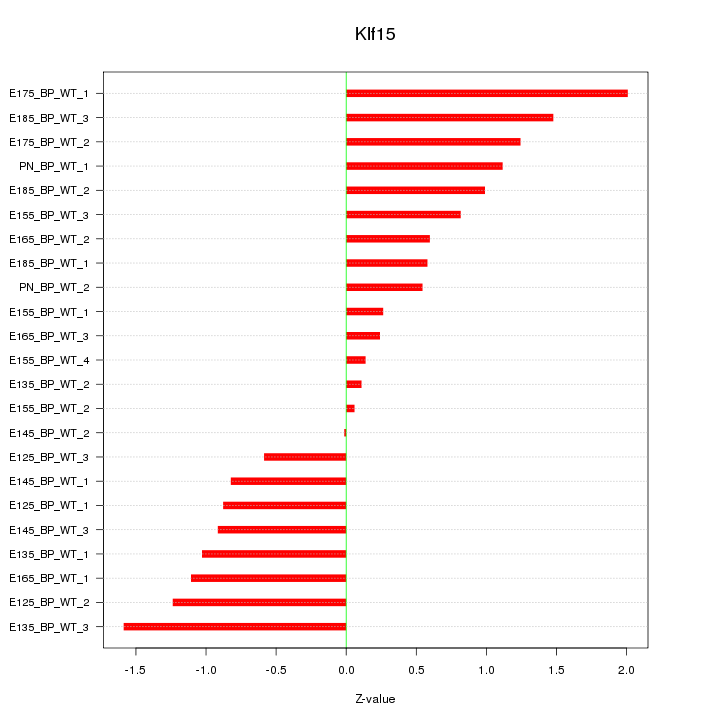
| 1.1 |
3.4 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 |
4.8 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.6 |
1.3 |
GO:0072103 |
glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.6 |
1.8 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.5 |
2.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 |
1.5 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.5 |
7.8 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.5 |
2.4 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.5 |
2.7 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 |
1.8 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 |
1.2 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.4 |
1.5 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.4 |
1.1 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.3 |
1.7 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.3 |
1.9 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 |
1.9 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.3 |
1.0 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 |
1.2 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.3 |
1.4 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 |
0.8 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.2 |
0.7 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.2 |
1.4 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 |
1.1 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
0.9 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 |
1.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.2 |
0.6 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 |
1.4 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.2 |
1.2 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
0.7 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 |
0.5 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 |
3.0 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.2 |
0.7 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 |
0.5 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.2 |
1.3 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
0.6 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.1 |
0.3 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.1 |
0.7 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
1.1 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 |
2.4 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.9 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.1 |
0.9 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
2.8 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.9 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) proline biosynthetic process(GO:0006561) |
| 0.1 |
1.5 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
1.1 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.1 |
0.6 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
1.0 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.7 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.6 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
1.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 |
0.4 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
0.4 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.6 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
1.3 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 |
1.4 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 |
0.7 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.4 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
2.0 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 |
1.1 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.3 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
1.6 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
1.2 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.3 |
GO:0035878 |
nail development(GO:0035878) |
| 0.1 |
1.8 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.1 |
1.7 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.5 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
1.6 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
1.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.4 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.4 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.1 |
0.8 |
GO:2000272 |
negative regulation of receptor activity(GO:2000272) |
| 0.0 |
0.8 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
1.0 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.9 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
1.0 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
1.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
1.0 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.5 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
1.2 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.4 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.0 |
0.2 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
1.3 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.3 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.9 |
GO:0032094 |
response to food(GO:0032094) |
| 0.0 |
0.0 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.6 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.6 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
1.2 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 |
1.0 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.4 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.4 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.3 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.5 |
GO:0061036 |
positive regulation of cartilage development(GO:0061036) |
| 0.0 |
2.6 |
GO:0006402 |
mRNA catabolic process(GO:0006402) |
| 0.0 |
0.4 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.0 |
1.5 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.1 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 |
0.1 |
GO:0010878 |
cholesterol storage(GO:0010878) |
| 0.0 |
0.3 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.1 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 |
1.3 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.6 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.7 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
1.3 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.2 |
GO:0060914 |
heart formation(GO:0060914) |
| 0.0 |
0.1 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.8 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.4 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.4 |
GO:0043297 |
apical junction assembly(GO:0043297) |
| 0.0 |
0.8 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
0.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.2 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.0 |
0.6 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
0.1 |
GO:0014053 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 |
0.2 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.0 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 |
0.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.3 |
GO:0007143 |
female meiotic division(GO:0007143) |