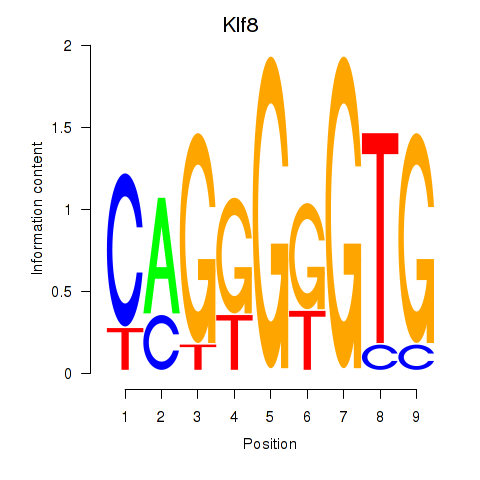
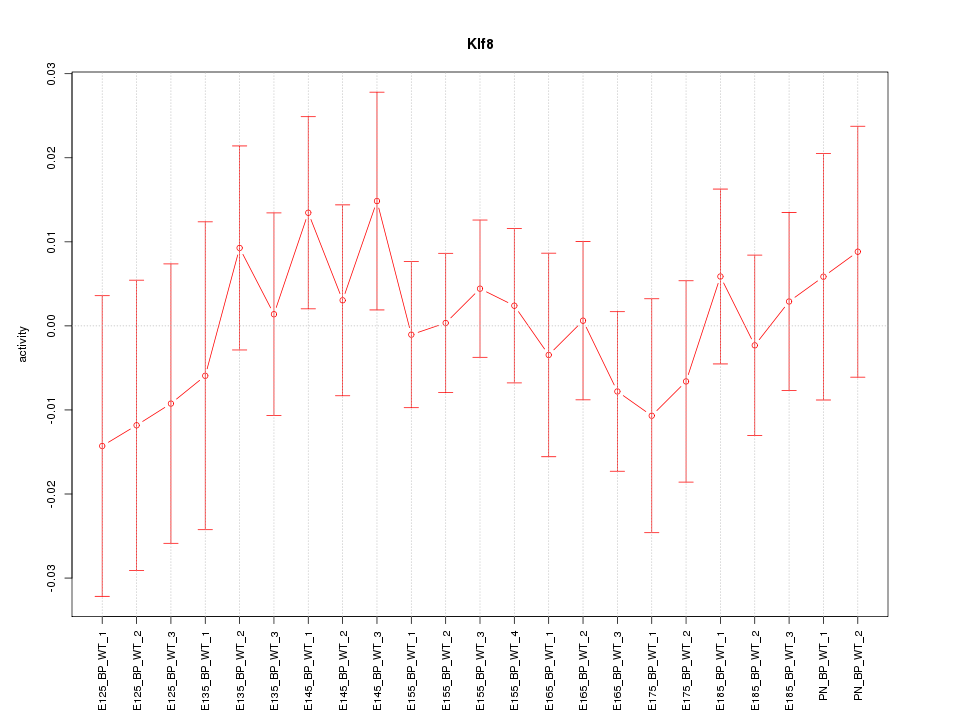
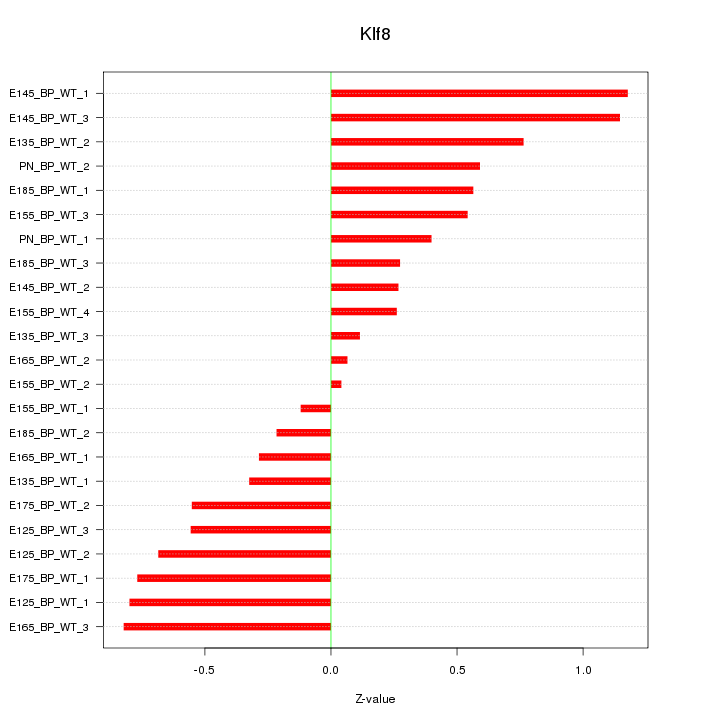
| 0.3 |
1.0 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 |
0.8 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 |
0.8 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 |
0.7 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 |
1.0 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.2 |
0.6 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 |
1.0 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 |
0.5 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.2 |
0.5 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.2 |
0.3 |
GO:1903225 |
regulation of endodermal cell differentiation(GO:1903224) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.7 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 |
0.7 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 |
1.0 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.2 |
GO:1905065 |
regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.1 |
0.7 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.5 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 |
0.3 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.5 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.4 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.3 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.1 |
0.6 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.4 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.1 |
0.5 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.3 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.6 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.3 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.6 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.3 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.4 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.5 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.1 |
GO:0061349 |
planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.1 |
0.2 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 |
0.1 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.9 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.4 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.1 |
0.4 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 |
0.2 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.1 |
0.2 |
GO:0046671 |
alpha-beta T cell lineage commitment(GO:0002363) positive regulation of neuron maturation(GO:0014042) regulation of cellular pH reduction(GO:0032847) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 |
0.2 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 |
0.2 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.2 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 |
0.1 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.0 |
0.6 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
1.0 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.1 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.0 |
0.1 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.5 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.4 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.2 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.3 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.0 |
0.3 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.4 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.4 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.3 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.3 |
GO:0042473 |
outer ear morphogenesis(GO:0042473) |
| 0.0 |
0.5 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
1.5 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.0 |
0.2 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 |
1.1 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
1.0 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.6 |
GO:0060384 |
innervation(GO:0060384) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.4 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.5 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.0 |
0.3 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.0 |
0.1 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
0.2 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.2 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.3 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.5 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.6 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.2 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.1 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.0 |
0.0 |
GO:0072338 |
creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) cellular lactam metabolic process(GO:0072338) |
| 0.0 |
0.2 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.0 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.0 |
0.2 |
GO:0036230 |
granulocyte activation(GO:0036230) |
| 0.0 |
0.2 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.0 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.2 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.2 |
GO:0045724 |
positive regulation of cilium assembly(GO:0045724) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.0 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.1 |
GO:0086064 |
cell communication by electrical coupling involved in cardiac conduction(GO:0086064) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.8 |
GO:0035383 |
acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |