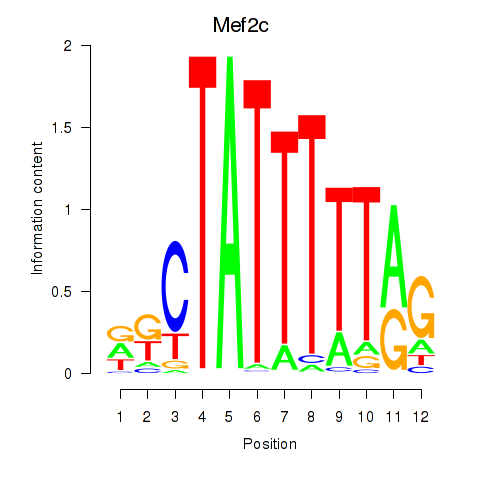
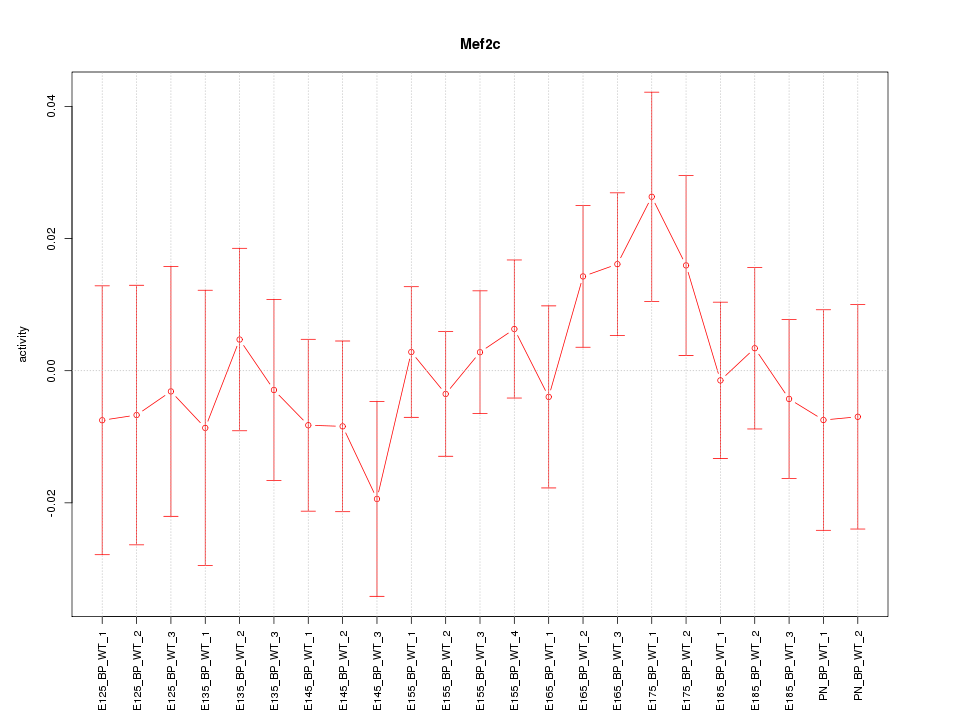
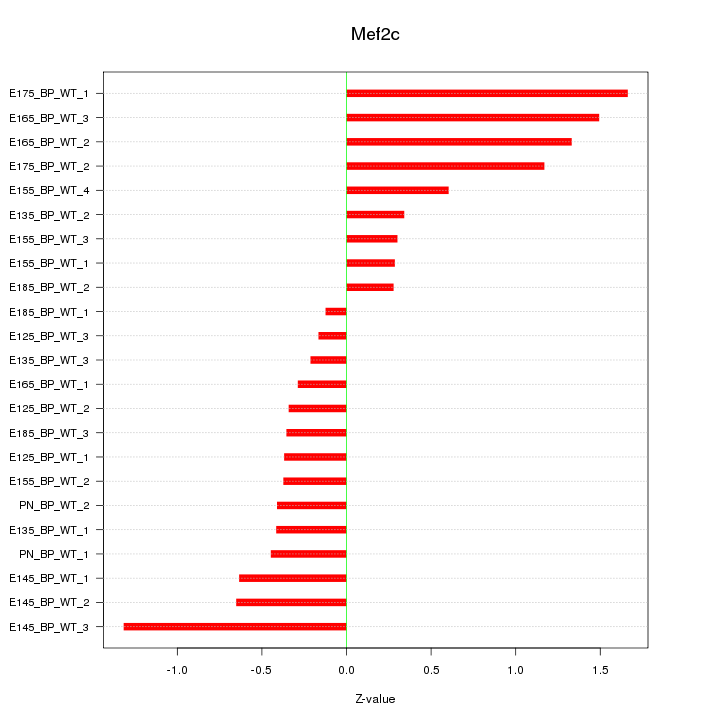
| 0.7 |
8.2 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.5 |
2.9 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 |
4.7 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 |
1.8 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.4 |
4.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 |
1.5 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.3 |
0.8 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 |
1.1 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.2 |
0.8 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 |
1.0 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
0.6 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.2 |
2.0 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 |
0.7 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.2 |
1.8 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 |
1.3 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.2 |
0.9 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
0.3 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
1.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.7 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 |
0.6 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.3 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
1.0 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
1.2 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 |
0.3 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.1 |
1.4 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.1 |
0.3 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 |
0.3 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
0.6 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.3 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 |
1.8 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.4 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 |
0.6 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
1.2 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.8 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.1 |
0.3 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
1.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
1.2 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.3 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.7 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.4 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.3 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0055118 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of cardiac muscle contraction(GO:0055118) negative regulation of mucus secretion(GO:0070256) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.2 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.0 |
0.4 |
GO:2000291 |
regulation of myoblast proliferation(GO:2000291) |
| 0.0 |
1.0 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.4 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.3 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.3 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 |
1.0 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.3 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
0.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.4 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.9 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.5 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
1.4 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
2.5 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.8 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |