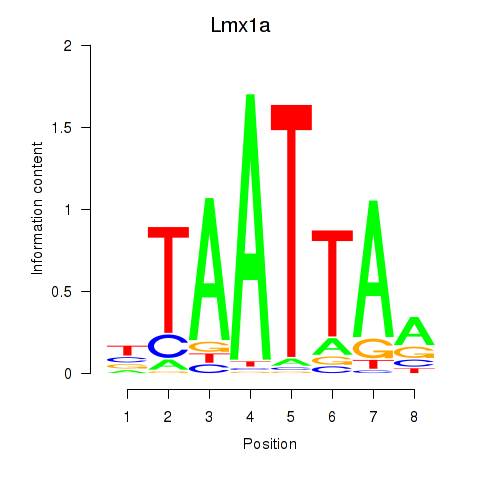
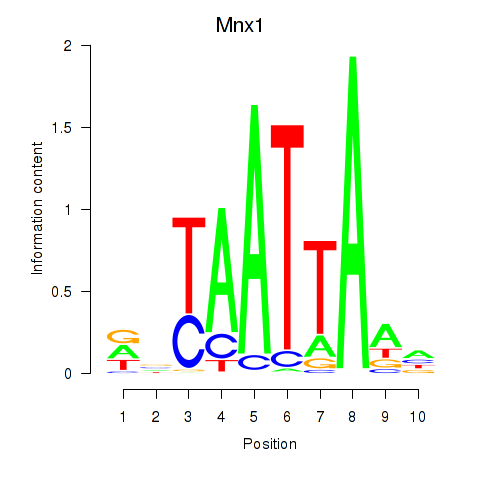
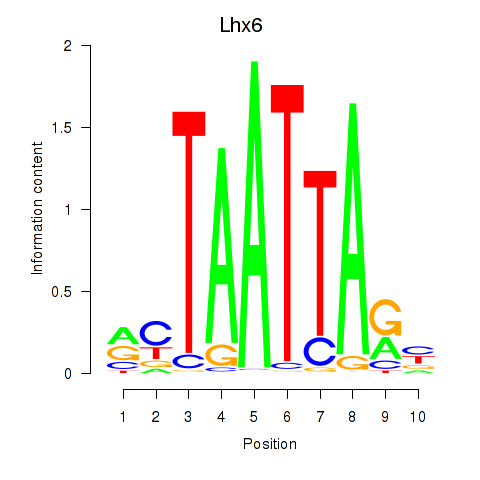
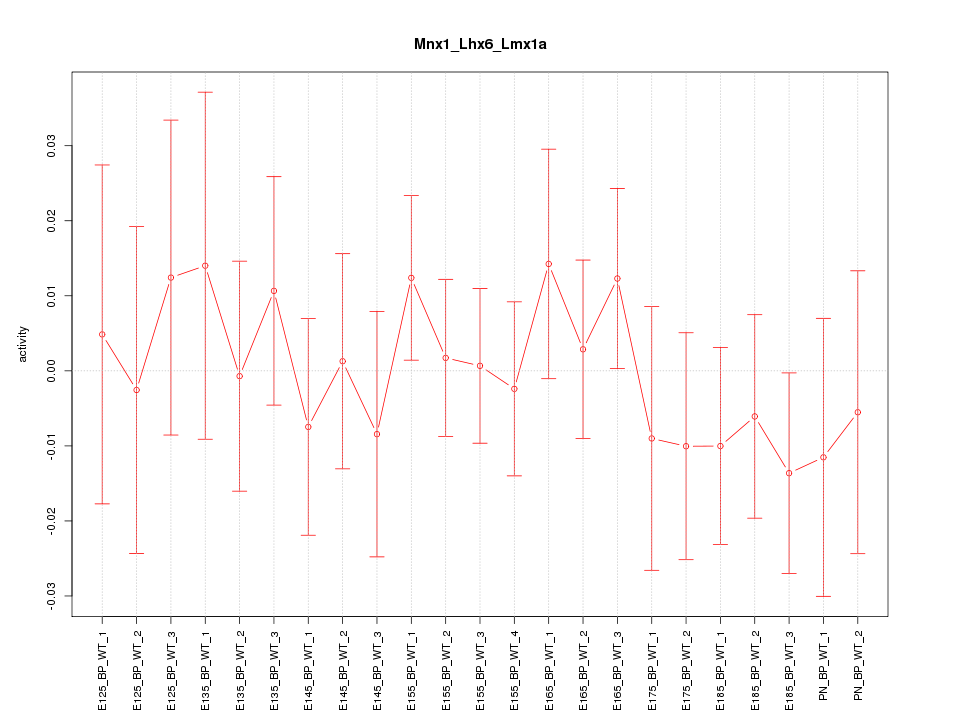
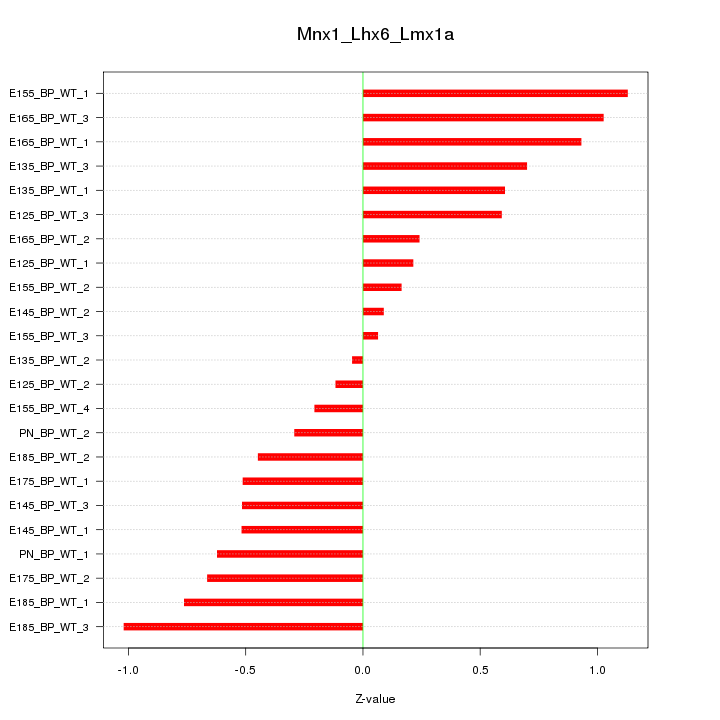
| 0.6 |
1.7 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 |
3.4 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 |
0.8 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 |
0.7 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.2 |
0.7 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.2 |
1.0 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.2 |
2.3 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.3 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.3 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
0.2 |
GO:0060523 |
prostate epithelial cord elongation(GO:0060523) |
| 0.1 |
1.1 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.9 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.7 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.1 |
0.3 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.2 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 |
1.3 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.5 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
0.2 |
GO:1905065 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.1 |
0.2 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
2.2 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.8 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.2 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.2 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.1 |
1.0 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.2 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
0.2 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.6 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.6 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.2 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
0.2 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
1.1 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.7 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.3 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.0 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.1 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.0 |
0.1 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 |
0.4 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.2 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
0.1 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 |
0.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.2 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.0 |
0.1 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.2 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.1 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.2 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 |
0.1 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.4 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.7 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.2 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.0 |
0.1 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.0 |
0.1 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.2 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 |
0.1 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 |
0.5 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.5 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.0 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 |
1.3 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.0 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.6 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 |
0.4 |
GO:0071384 |
cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.7 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.8 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 |
0.1 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.1 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.0 |
0.2 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
2.9 |
GO:0043010 |
camera-type eye development(GO:0043010) |
| 0.0 |
0.1 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |