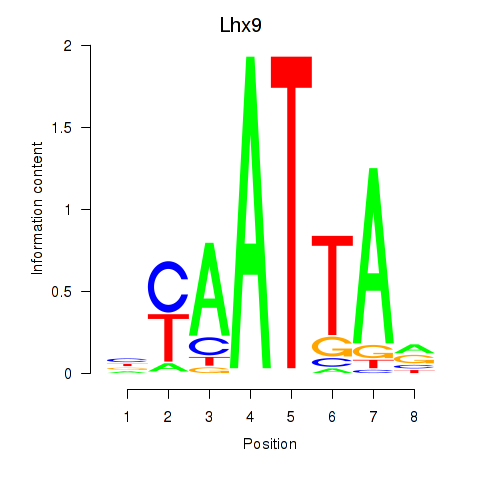
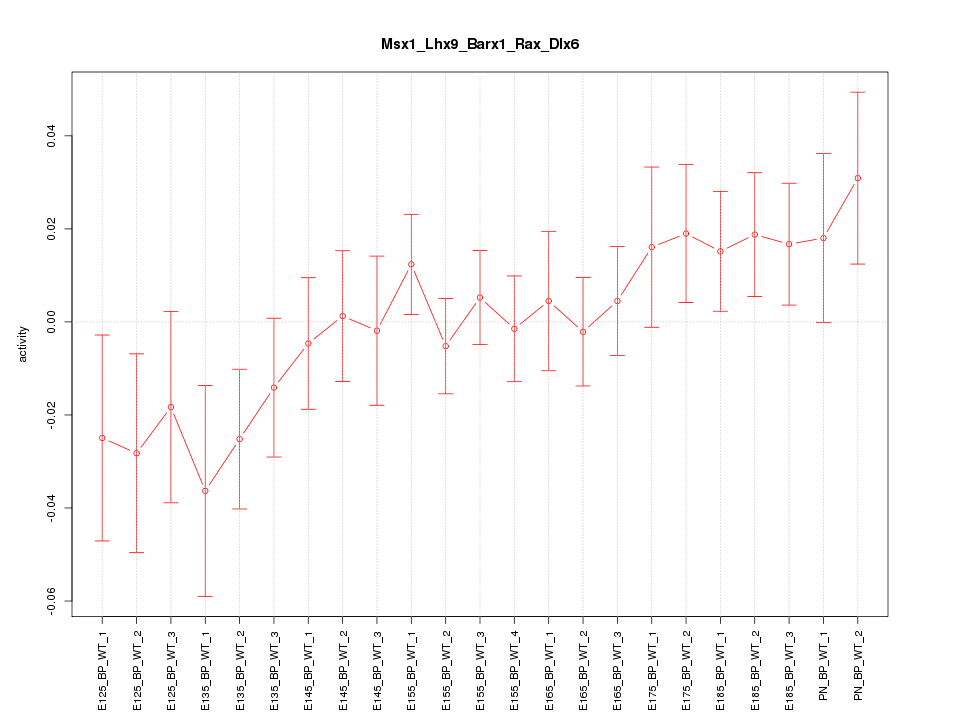
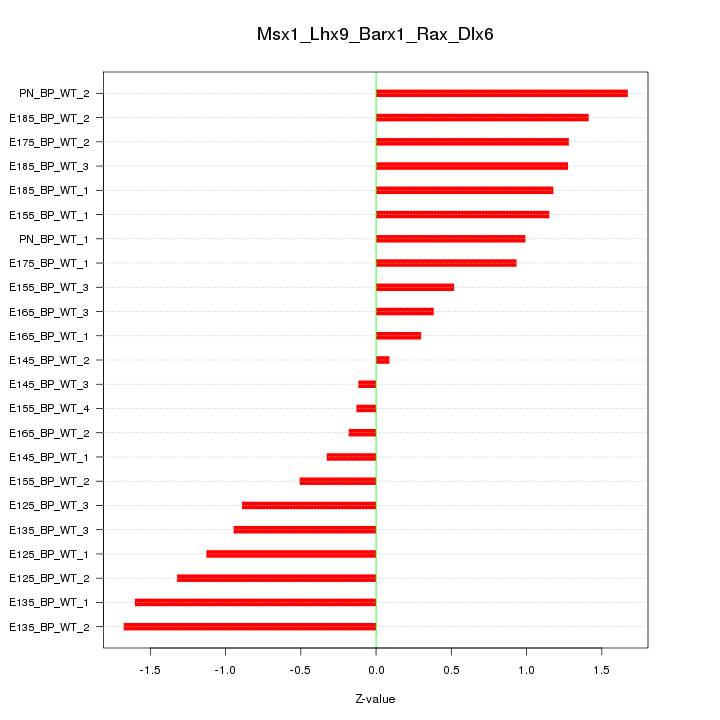
| 1.8 |
1.8 |
GO:0060915 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 1.6 |
4.8 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.6 |
4.7 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.8 |
3.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.8 |
3.9 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.8 |
5.4 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.7 |
2.0 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.6 |
5.6 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.6 |
3.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 |
10.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 |
1.7 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.4 |
1.2 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.3 |
1.7 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 |
0.9 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 |
8.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.2 |
1.5 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.2 |
0.9 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 |
0.3 |
GO:0048041 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.2 |
1.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
0.8 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.2 |
7.5 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.2 |
3.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
1.3 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 |
0.4 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.4 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.1 |
0.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
1.7 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
1.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.1 |
13.8 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 |
0.9 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 |
2.0 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.6 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
0.3 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 |
0.7 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.6 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.4 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.4 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.2 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 |
1.1 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.2 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.2 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.2 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.1 |
0.4 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.8 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 |
1.4 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.1 |
0.4 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
1.5 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.1 |
0.4 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
1.6 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.6 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
1.2 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.7 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.7 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
5.0 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
1.1 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
4.6 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.7 |
GO:1904892 |
regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 |
3.1 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.7 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
3.1 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.0 |
2.3 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.7 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.2 |
GO:0034397 |
telomere localization(GO:0034397) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.7 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
1.6 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.0 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.0 |
0.2 |
GO:0035886 |
vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 |
0.1 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.2 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.0 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.4 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |