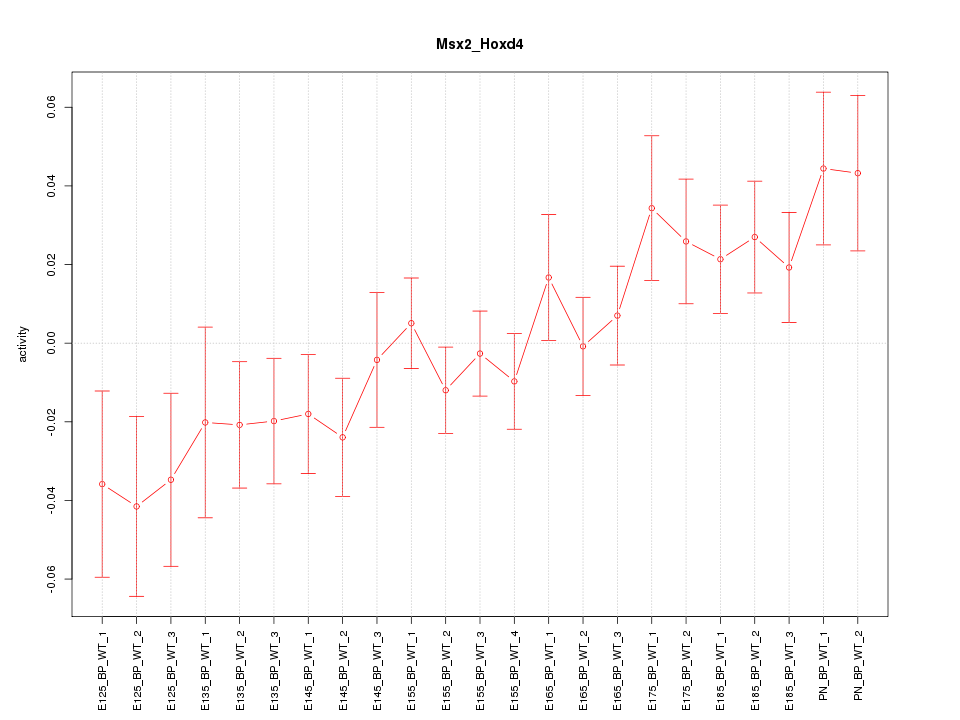
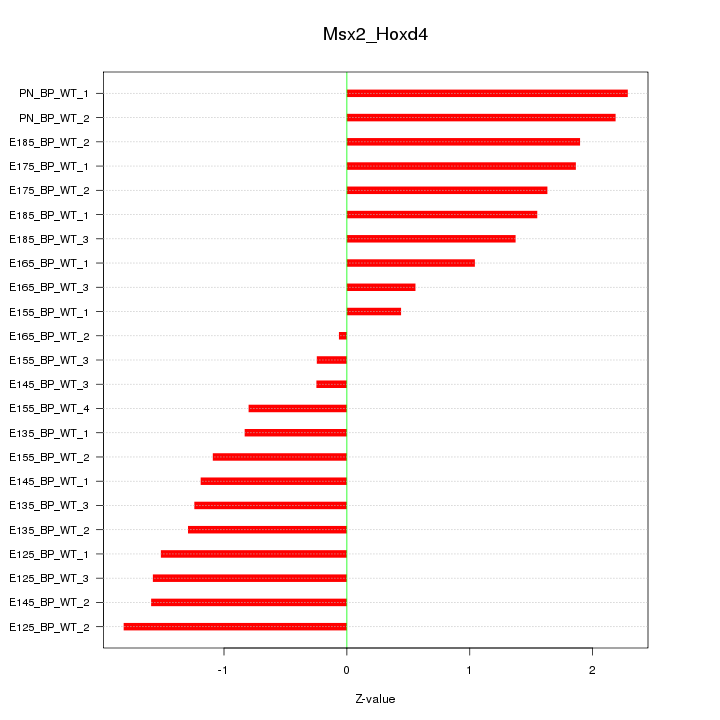
Motif ID: Msx2_Hoxd4
Z-value: 1.376
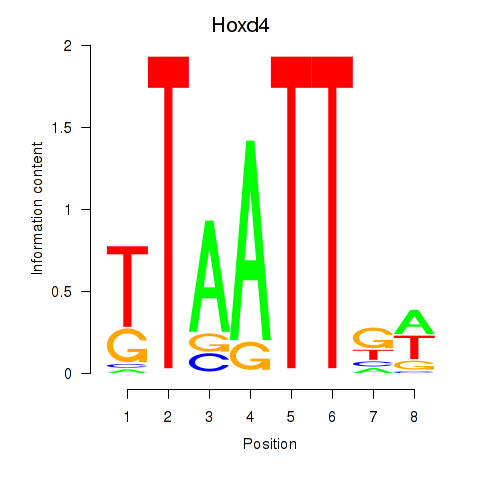
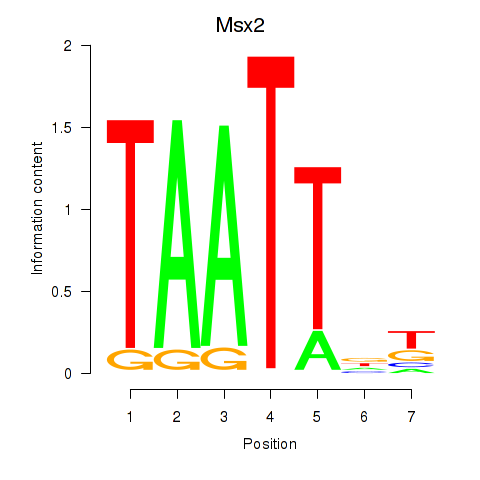
Transcription factors associated with Msx2_Hoxd4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Msx2 | ENSMUSG00000021469.8 | Msx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | mm10_v2_chr13_-_53473074_53473074 | -0.11 | 6.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.8 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 2.9 | 8.8 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 | 1.7 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.5 | 1.4 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.5 | 1.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.3 | 6.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 0.9 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.3 | 0.9 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 6.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 1.8 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 0.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.5 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 0.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 2.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 2.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.7 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.2 | 2.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 1.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 5.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 5.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.9 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.9 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 2.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 1.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 6.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 2.0 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 11.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 2.2 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 1.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.7 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 4.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.7 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.7 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.6 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.4 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 2.4 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 1.0 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.6 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.5 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 1.4 | GO:0015031 | protein transport(GO:0015031) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 1.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.3 | 2.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 0.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 1.7 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 8.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 18.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.7 | 21.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.6 | 1.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.6 | 11.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.5 | 1.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 14.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 1.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.0 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 5.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 2.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 5.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 4.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 12.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.7 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |