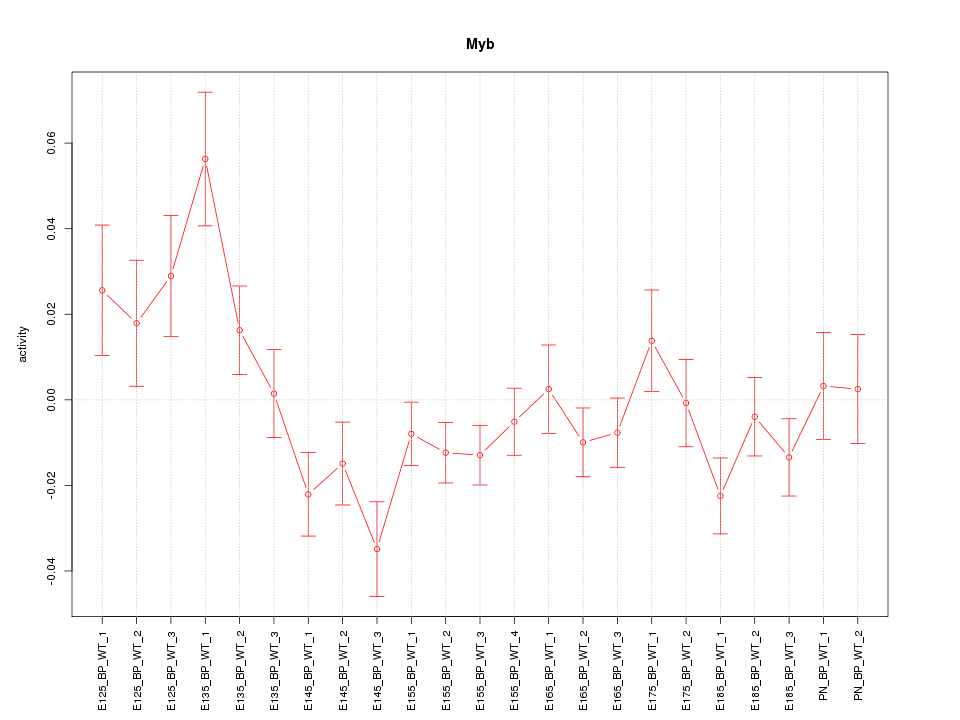
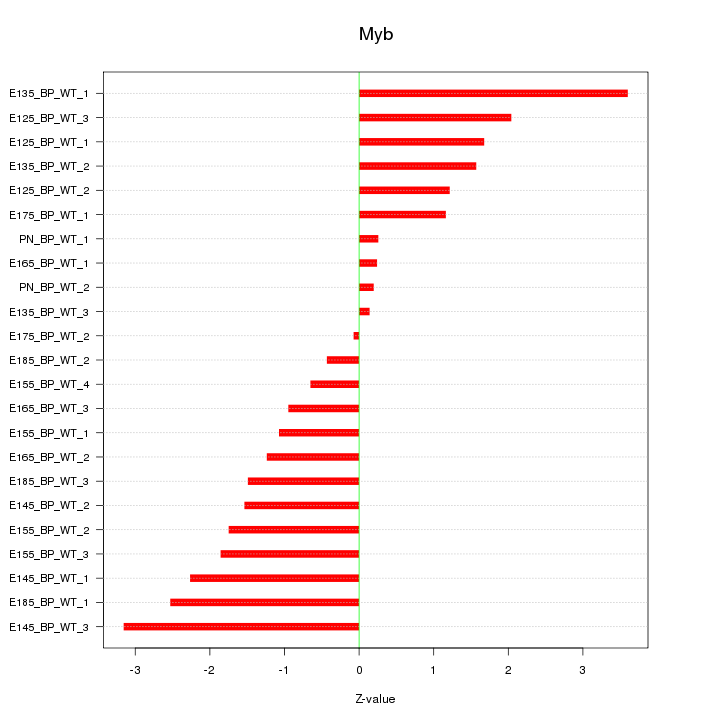
Motif ID: Myb
Z-value: 1.645
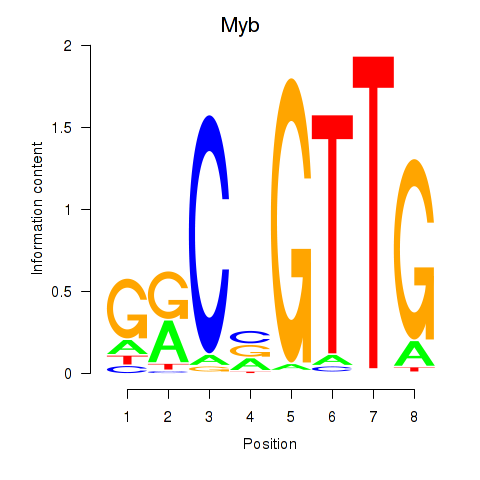
Transcription factors associated with Myb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Myb | ENSMUSG00000019982.8 | Myb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | mm10_v2_chr10_-_21160925_21160984 | 0.73 | 7.6e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.8 | 4.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.6 | 2.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.6 | 5.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.6 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 9.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 | 2.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.5 | 4.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 1.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 3.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.5 | 1.4 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.4 | 2.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 | 2.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 7.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.4 | 2.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.7 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 1.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.4 | 1.1 | GO:1901420 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) negative regulation of response to alcohol(GO:1901420) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.4 | 6.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.4 | 2.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.3 | 1.7 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) response to glucagon(GO:0033762) |
| 0.3 | 1.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.3 | 1.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 3.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 0.8 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 | 1.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.3 | 5.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 4.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 3.8 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.2 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.8 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.8 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 2.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 3.4 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.2 | 5.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 5.2 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 2.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 2.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.4 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 3.6 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 3.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.2 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.5 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.5 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 1.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) regulation of determination of dorsal identity(GO:2000015) |
| 0.1 | 1.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 1.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 0.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 1.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 0.9 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.5 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 3.0 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.3 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.7 | GO:0060049 | protein O-linked mannosylation(GO:0035269) regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 1.5 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 5.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.2 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.9 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.1 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 2.1 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.6 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.9 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 3.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.1 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 6.1 | GO:0007067 | mitotic nuclear division(GO:0007067) |
| 0.0 | 1.6 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.3 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 2.5 | GO:0044772 | mitotic cell cycle phase transition(GO:0044772) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 2.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.1 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.6 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 3.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.6 | 1.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.5 | 1.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 3.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 8.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 0.8 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.3 | 1.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 1.6 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.2 | 4.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 6.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 3.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 0.9 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 2.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 0.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 3.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 2.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 7.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 5.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 11.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 5.9 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 3.0 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.2 | 5.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.6 | 2.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 3.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 3.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.5 | 3.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.5 | 1.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.4 | 5.4 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.4 | 2.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 1.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 1.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 1.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.3 | 7.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 0.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 1.5 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.2 | 1.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 3.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.8 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.7 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.1 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 3.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 7.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 3.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 3.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 2.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.9 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 5.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 5.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |