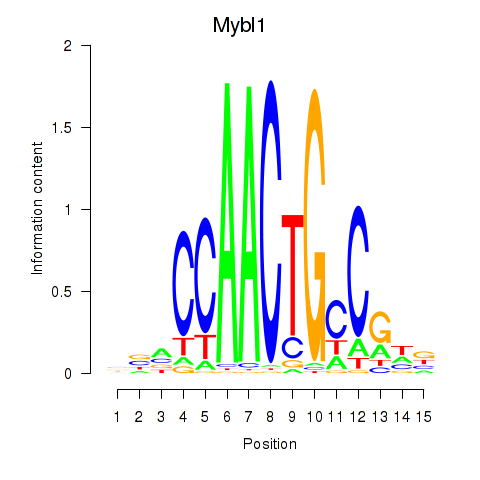
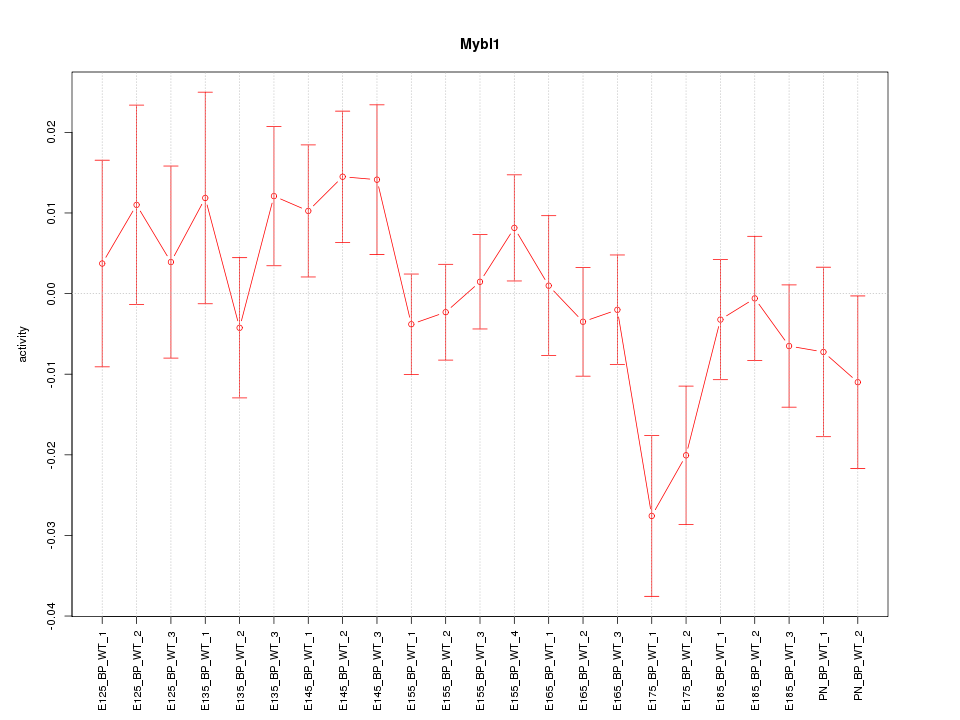
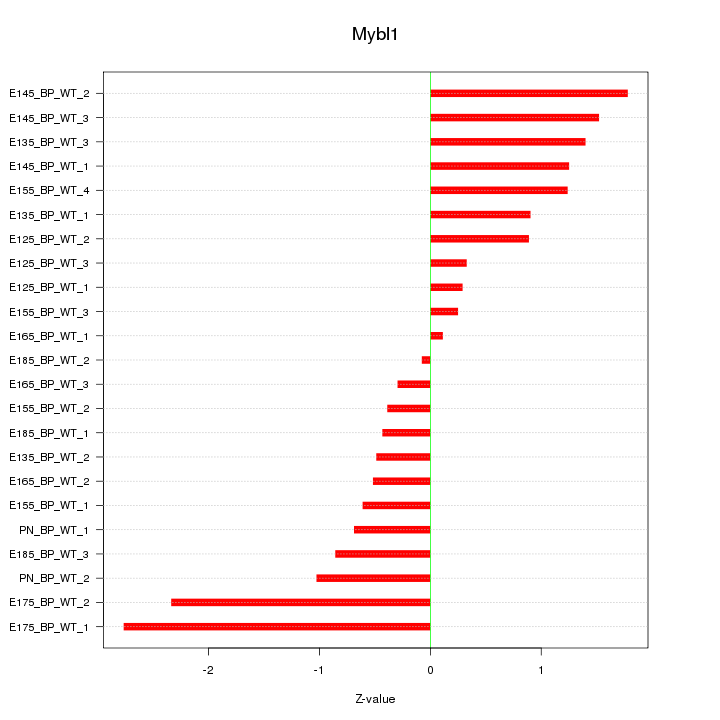
| 1.0 |
6.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.9 |
3.5 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.5 |
1.5 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 |
1.8 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.4 |
2.2 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.4 |
1.2 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 |
1.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.3 |
1.7 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
4.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.3 |
2.9 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.3 |
0.9 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.3 |
1.5 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.3 |
0.8 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.3 |
2.1 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.3 |
1.0 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.3 |
2.3 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 |
0.8 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 |
1.3 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 |
0.7 |
GO:1902174 |
striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 |
5.7 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 |
0.4 |
GO:1901740 |
negative regulation of myoblast fusion(GO:1901740) |
| 0.2 |
0.6 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 |
3.8 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 |
0.6 |
GO:0040009 |
regulation of growth rate(GO:0040009) protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 |
1.0 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.2 |
0.6 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 |
0.8 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 |
1.0 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 |
1.0 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 |
0.6 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.9 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.2 |
0.9 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.2 |
0.4 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 |
0.9 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.2 |
0.5 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 |
0.5 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.2 |
0.5 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
0.8 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
0.5 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.2 |
0.6 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.2 |
0.3 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.2 |
0.5 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.6 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.3 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.7 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.6 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 |
0.4 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.5 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
1.0 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
1.2 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.1 |
0.4 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 |
0.7 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
1.6 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
0.7 |
GO:0090666 |
telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
0.4 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.5 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
1.0 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.4 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.7 |
GO:0035087 |
siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 |
0.4 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 |
0.4 |
GO:2000334 |
response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 |
0.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
0.6 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
1.5 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.1 |
1.8 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.1 |
0.3 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
1.4 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.4 |
GO:0046709 |
IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
1.8 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.1 |
0.6 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.6 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 |
0.4 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.4 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.8 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
1.1 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 |
0.3 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.3 |
GO:0019405 |
hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.1 |
0.3 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.4 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.6 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.4 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.1 |
0.6 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.7 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
0.2 |
GO:1902036 |
regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 |
0.4 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 |
0.4 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
3.7 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.1 |
0.8 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
2.2 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.2 |
GO:0010886 |
positive regulation of cholesterol storage(GO:0010886) |
| 0.1 |
1.0 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 |
0.3 |
GO:0071034 |
CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.2 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.1 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.4 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
0.5 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.1 |
0.3 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
0.5 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 |
0.9 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.6 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
1.1 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.5 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.3 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 |
0.1 |
GO:0032714 |
negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 |
0.2 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 |
1.1 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.3 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.1 |
0.6 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.2 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
0.2 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.5 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.2 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 |
0.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.4 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.4 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.1 |
0.4 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
0.8 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.1 |
0.9 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.3 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.1 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 |
1.0 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.7 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.3 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
3.1 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.3 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.3 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
1.0 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.5 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.0 |
0.2 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.2 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.0 |
0.3 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.1 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 |
1.2 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
1.0 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.3 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.1 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.2 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.0 |
0.7 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.3 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.5 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
1.0 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
1.0 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.2 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.8 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.2 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.3 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.4 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
1.0 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.5 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.3 |
GO:0002155 |
regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 |
0.5 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.4 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.4 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.4 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
1.4 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.1 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 |
1.2 |
GO:0070303 |
negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 |
0.3 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.3 |
GO:0046929 |
negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 |
0.5 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.8 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.7 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
1.0 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.6 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.4 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.3 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.1 |
GO:0006534 |
cysteine metabolic process(GO:0006534) |
| 0.0 |
1.1 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.3 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.6 |
GO:0033108 |
mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.2 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
1.5 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.0 |
0.2 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.0 |
0.1 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.1 |
GO:0032201 |
telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 |
0.1 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 |
0.7 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.1 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.9 |
GO:0008637 |
apoptotic mitochondrial changes(GO:0008637) |
| 0.0 |
0.4 |
GO:0042417 |
dopamine metabolic process(GO:0042417) |
| 0.0 |
0.3 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.2 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) |
| 0.0 |
0.0 |
GO:2000620 |
regulation of histone H4-K16 acetylation(GO:2000618) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.0 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 |
0.1 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
0.4 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.1 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.0 |
0.2 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.3 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.4 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.2 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.2 |
GO:0009651 |
response to salt stress(GO:0009651) |
| 0.0 |
0.3 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.3 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.1 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 |
0.5 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.2 |
GO:0030220 |
platelet formation(GO:0030220) |
| 0.0 |
0.3 |
GO:1904407 |
positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 |
1.4 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.3 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.2 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 |
0.3 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.1 |
GO:0010455 |
negative regulation of cell fate commitment(GO:0010454) positive regulation of cell fate commitment(GO:0010455) regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.4 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) |
| 0.0 |
0.2 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.2 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.5 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |