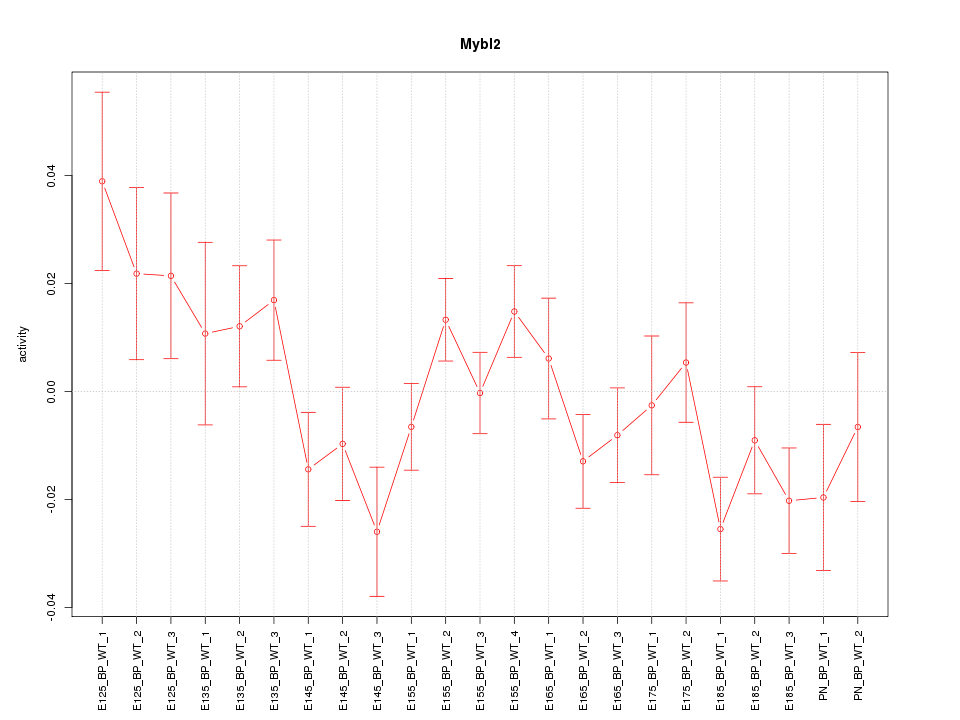
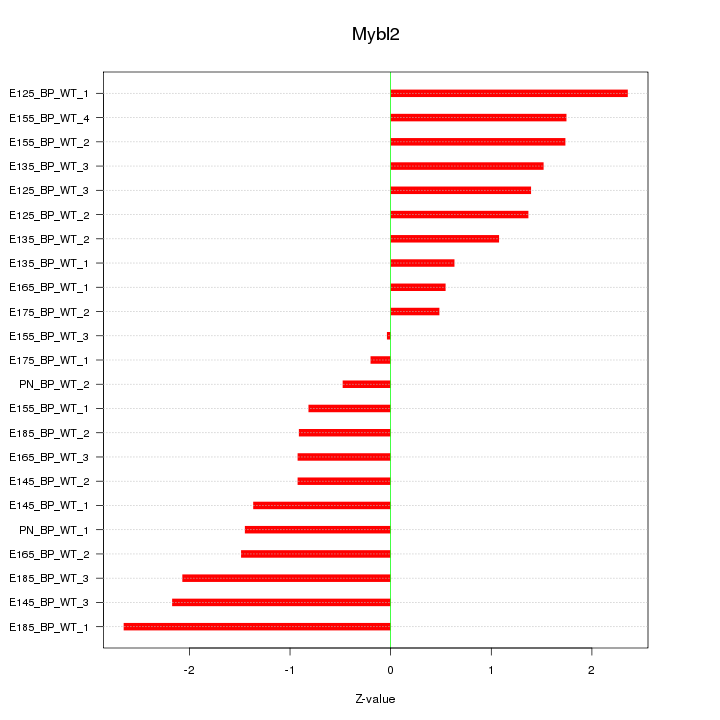
Motif ID: Mybl2
Z-value: 1.407
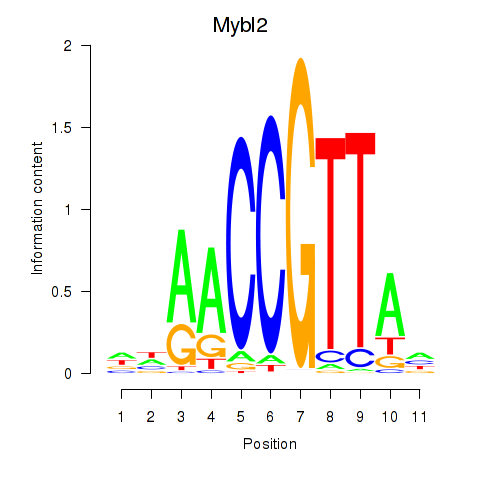
Transcription factors associated with Mybl2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mybl2 | ENSMUSG00000017861.5 | Mybl2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl2 | mm10_v2_chr2_+_163054682_163054693 | 0.49 | 1.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.7 | 2.0 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.6 | 1.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.6 | 2.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.4 | 1.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 3.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 1.2 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.4 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.4 | 1.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 | 0.8 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.3 | 1.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.3 | 0.8 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 2.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 1.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 1.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 4.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 5.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.6 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 1.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.9 | GO:0098532 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 0.7 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 5.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 0.8 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.2 | 2.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 2.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 0.9 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 1.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 2.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 2.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.9 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 1.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 2.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.8 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.9 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 0.3 | GO:0046655 | glycine biosynthetic process(GO:0006545) folic acid metabolic process(GO:0046655) |
| 0.1 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.4 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 3.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 2.6 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.4 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0045842 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.9 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 2.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.5 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.1 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0097343 | positive regulation of T cell receptor signaling pathway(GO:0050862) ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.7 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.6 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.0 | 6.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.8 | 2.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 1.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 1.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.3 | 1.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 0.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 0.8 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 2.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 1.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 2.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 7.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.3 | GO:0071204 | CRD-mediated mRNA stability complex(GO:0070937) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 2.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 2.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 3.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 1.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 1.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.4 | 1.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.3 | 2.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 2.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.3 | 0.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.3 | 0.8 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.3 | 0.8 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.2 | 1.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 5.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 2.0 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.2 | 1.0 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 5.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.1 | GO:0017161 | JUN kinase phosphatase activity(GO:0008579) phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 3.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0070181 | mRNA 5'-UTR binding(GO:0048027) small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 1.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.0 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 1.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.6 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |