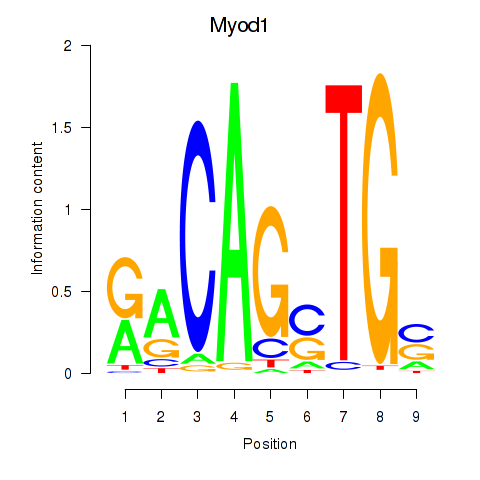
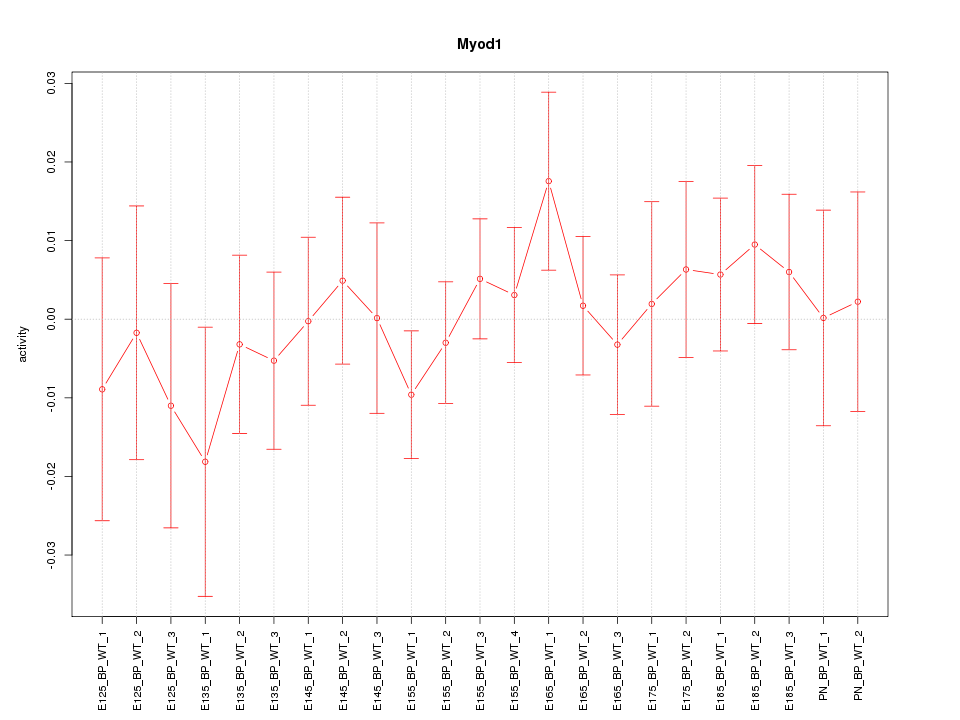
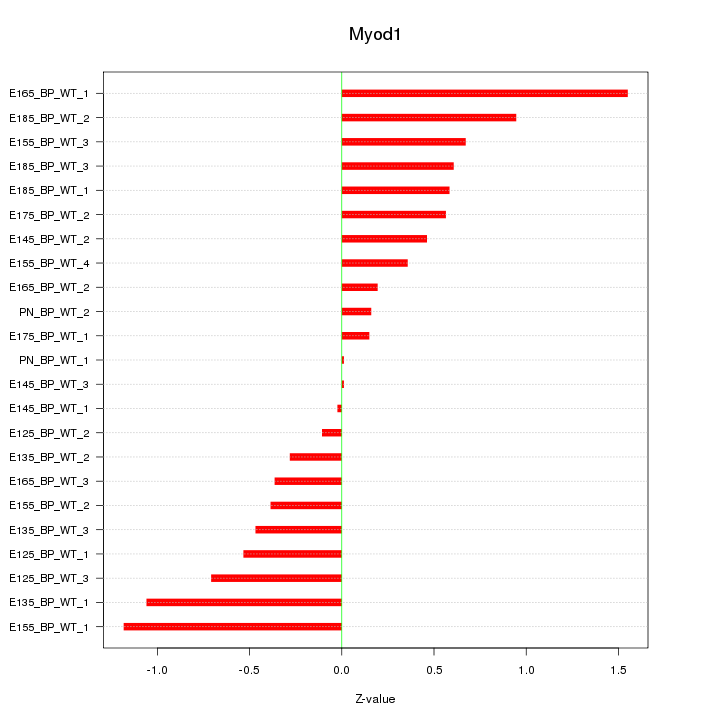
| 0.5 |
1.6 |
GO:0051464 |
positive regulation of cortisol secretion(GO:0051464) |
| 0.5 |
2.0 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.4 |
1.3 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.3 |
1.0 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 |
2.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.3 |
2.3 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 |
1.0 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 |
0.9 |
GO:0046959 |
habituation(GO:0046959) |
| 0.3 |
1.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 |
0.6 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 |
1.1 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.3 |
1.6 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 |
1.0 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
0.7 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 |
1.8 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 |
1.4 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.2 |
0.7 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.2 |
2.2 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.6 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
0.6 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.2 |
0.6 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.2 |
0.6 |
GO:0061324 |
canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.2 |
1.0 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 |
0.9 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.2 |
2.1 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 |
1.2 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 |
0.4 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.1 |
0.4 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
0.4 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 |
0.6 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.6 |
GO:2001137 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.4 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
0.3 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.5 |
GO:0003430 |
tolerance induction to self antigen(GO:0002513) growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
0.6 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
1.3 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.1 |
1.3 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.4 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.1 |
0.6 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.4 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 |
0.9 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.7 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.3 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.3 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
1.1 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.5 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
1.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 |
0.9 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 |
0.5 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.1 |
0.5 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.7 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.8 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.1 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
0.5 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.5 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 |
0.7 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.3 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.1 |
0.2 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 |
0.5 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
0.7 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 |
0.3 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.4 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.3 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 |
0.2 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
1.1 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.4 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
1.1 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.2 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 |
0.5 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.0 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.2 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.1 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.0 |
1.7 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
0.2 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 |
0.3 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
0.3 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.1 |
GO:0002586 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 |
0.8 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.5 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.5 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
1.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.9 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.3 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.5 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.4 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 |
0.5 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.5 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.0 |
1.0 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.8 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.3 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.4 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.4 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.1 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.7 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
1.6 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.3 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
1.3 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.0 |
GO:0061502 |
early endosome to recycling endosome transport(GO:0061502) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.2 |
GO:1902170 |
cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 |
0.5 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.0 |
0.4 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.3 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.5 |
GO:0060749 |
mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 |
0.8 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.1 |
GO:0010455 |
positive regulation of cell fate commitment(GO:0010455) |
| 0.0 |
0.1 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 |
0.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.1 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 |
0.2 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.2 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.2 |
GO:0045634 |
regulation of melanocyte differentiation(GO:0045634) |
| 0.0 |
0.3 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.4 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.0 |
0.3 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 |
0.6 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.7 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0060979 |
vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 |
0.1 |
GO:1903887 |
common bile duct development(GO:0061009) motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.1 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.0 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.2 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.1 |
GO:0035509 |
negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 |
0.0 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 |
0.2 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.2 |
GO:0015012 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.3 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.2 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
1.0 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.7 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
1.1 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.0 |
1.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.0 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.0 |
0.3 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.9 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
0.1 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.1 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.0 |
0.0 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.0 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.1 |
GO:0019363 |
pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 |
0.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.4 |
GO:0042073 |
intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |