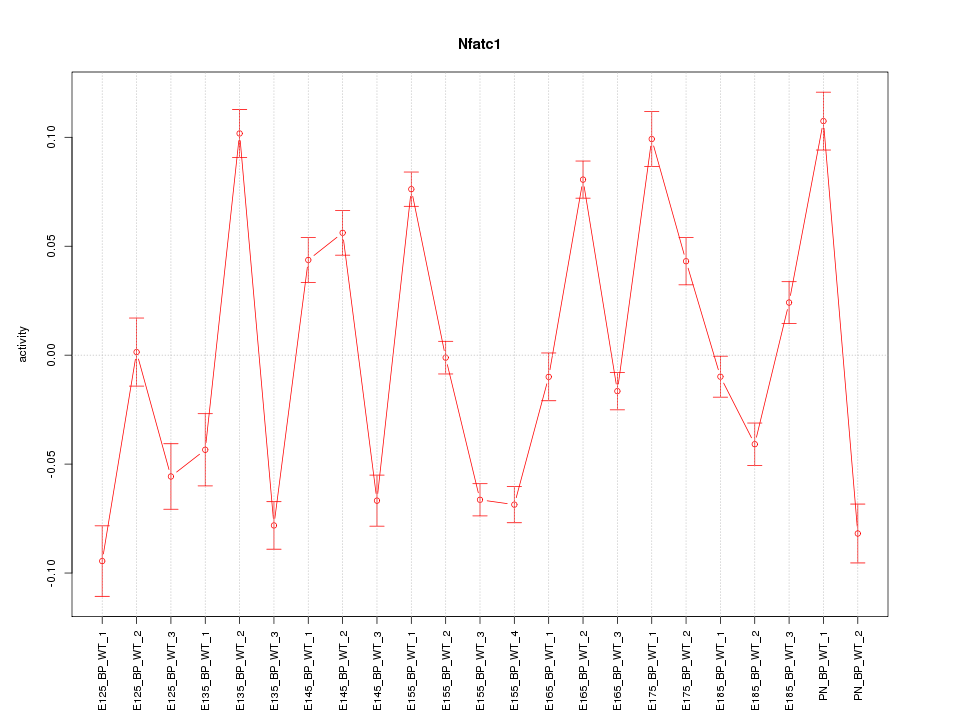
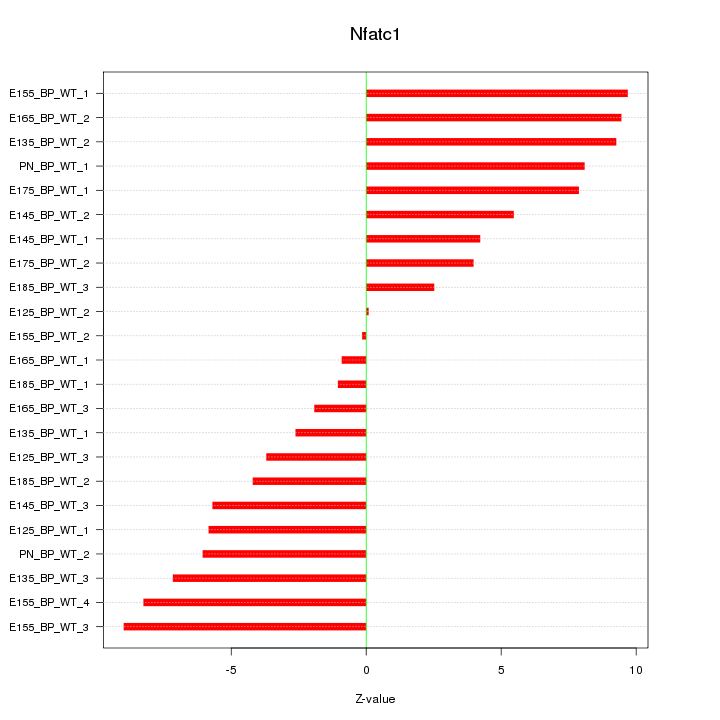
Motif ID: Nfatc1
Z-value: 5.949
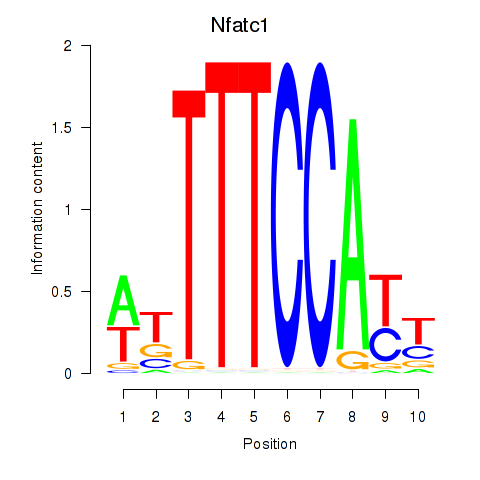
Transcription factors associated with Nfatc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc1 | ENSMUSG00000033016.9 | Nfatc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80708115_80708180 | -0.18 | 4.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 3.1 | 12.3 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 2.4 | 7.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 2.3 | 7.0 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 2.2 | 10.9 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 2.0 | 8.2 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.6 | 4.7 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 1.5 | 9.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.4 | 10.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.9 | 6.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.9 | 8.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.9 | 7.7 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.8 | 2.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.8 | 2.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.8 | 3.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.7 | 3.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.6 | 1.9 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.6 | 2.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.6 | 2.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 1.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 | 4.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 2.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.4 | 2.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.4 | 0.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.4 | 2.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.3 | 3.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 3.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.3 | 3.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 4.0 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 0.9 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 3.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 0.9 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.3 | 1.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 1.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 3.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 2.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 6.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 2.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 3.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 1.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 4.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 | 2.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.7 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 4.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.5 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.9 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 2.0 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.4 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 2.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.8 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 669.7 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.1 | 12.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.9 | 3.6 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 3.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.6 | 12.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 10.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 4.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 0.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 2.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 2.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 2.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 6.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 3.2 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 3.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 4.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 796.9 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.2 | 8.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.2 | 4.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.0 | 7.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.9 | 3.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 10.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 1.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 3.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 1.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 2.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.4 | 14.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 2.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 0.9 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 3.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 4.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 2.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.2 | 4.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 6.7 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 7.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.2 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 2.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 2.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 3.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.5 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 4.0 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 769.4 | GO:0003674 | molecular_function(GO:0003674) |