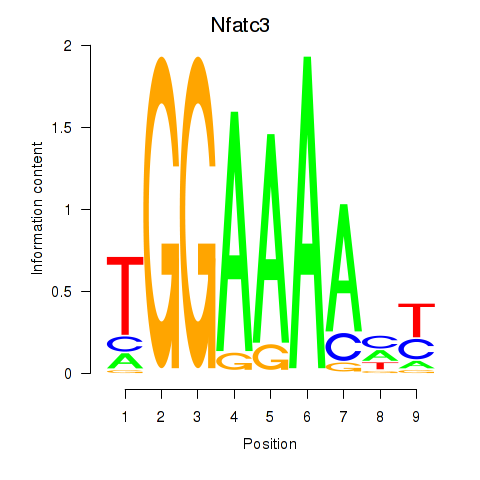
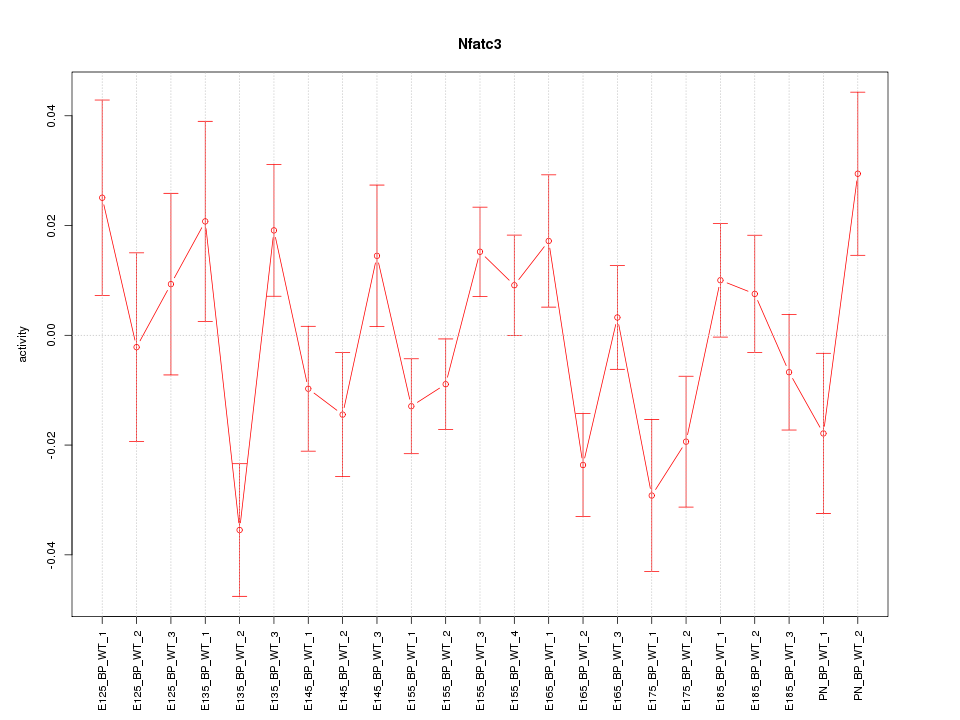
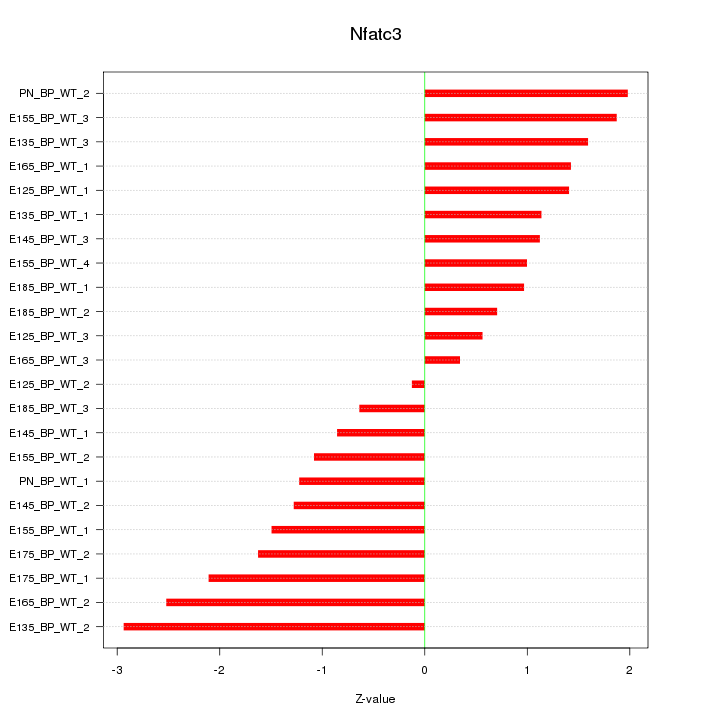
| 1.0 |
5.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 |
2.6 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.7 |
4.9 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.7 |
2.8 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.7 |
2.0 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.6 |
1.7 |
GO:1903903 |
striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.6 |
1.7 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.5 |
2.5 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.4 |
1.2 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 |
1.6 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.4 |
2.0 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 |
1.3 |
GO:0010749 |
regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.3 |
1.0 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 |
1.6 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 |
2.2 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.3 |
0.6 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.3 |
0.8 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 |
1.1 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
0.7 |
GO:0033092 |
positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 |
0.7 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 |
1.8 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 |
0.7 |
GO:2000977 |
oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.2 |
0.7 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 |
1.1 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 |
0.6 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 |
3.6 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
1.4 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 |
1.3 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 |
1.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 |
0.5 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 |
0.8 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.2 |
0.5 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 |
1.8 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 |
2.8 |
GO:0060579 |
ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.1 |
1.2 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 |
1.7 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.5 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.1 |
0.5 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 |
1.0 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.1 |
1.9 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.7 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.8 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.1 |
0.5 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.9 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
1.0 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.1 |
0.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.5 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.6 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.1 |
0.4 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.1 |
0.7 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.3 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.1 |
0.9 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.3 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.3 |
GO:0070543 |
response to linoleic acid(GO:0070543) |
| 0.1 |
0.6 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.4 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) |
| 0.1 |
1.4 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.1 |
0.2 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.3 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 |
0.5 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 |
0.7 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.3 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.2 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.1 |
0.4 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
1.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
0.7 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.8 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.7 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
0.9 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.4 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.7 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
0.3 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 |
0.3 |
GO:0090240 |
positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 |
0.8 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.1 |
0.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.1 |
0.4 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.7 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.8 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.1 |
0.3 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.3 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.2 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
1.1 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.2 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.3 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.6 |
GO:2000821 |
synaptic vesicle maturation(GO:0016188) AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.4 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 |
0.2 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.4 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) positive regulation of protein depolymerization(GO:1901881) |
| 0.0 |
0.1 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.0 |
1.0 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.3 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.0 |
0.1 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.2 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.0 |
0.1 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.3 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.2 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 |
0.4 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
0.1 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.0 |
0.6 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.2 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.8 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.2 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.1 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.0 |
0.5 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.4 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.5 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.8 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
2.9 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.9 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.1 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.0 |
0.1 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.0 |
0.3 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.1 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.0 |
0.5 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.1 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.6 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.3 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.1 |
GO:0071711 |
basement membrane organization(GO:0071711) |
| 0.0 |
0.1 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 |
0.6 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
2.4 |
GO:0048754 |
branching morphogenesis of an epithelial tube(GO:0048754) |
| 0.0 |
0.1 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 |
0.4 |
GO:0021904 |
dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
1.1 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |
| 0.0 |
0.4 |
GO:0042026 |
protein refolding(GO:0042026) |
| 0.0 |
0.3 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.4 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.1 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.2 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.3 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0010883 |
regulation of lipid storage(GO:0010883) |