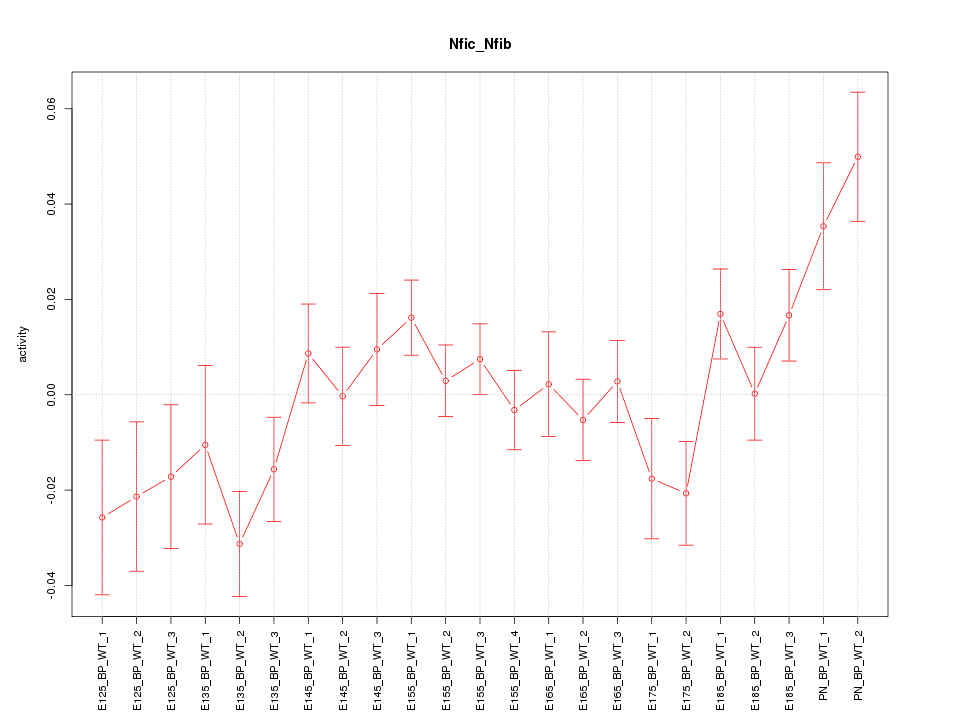
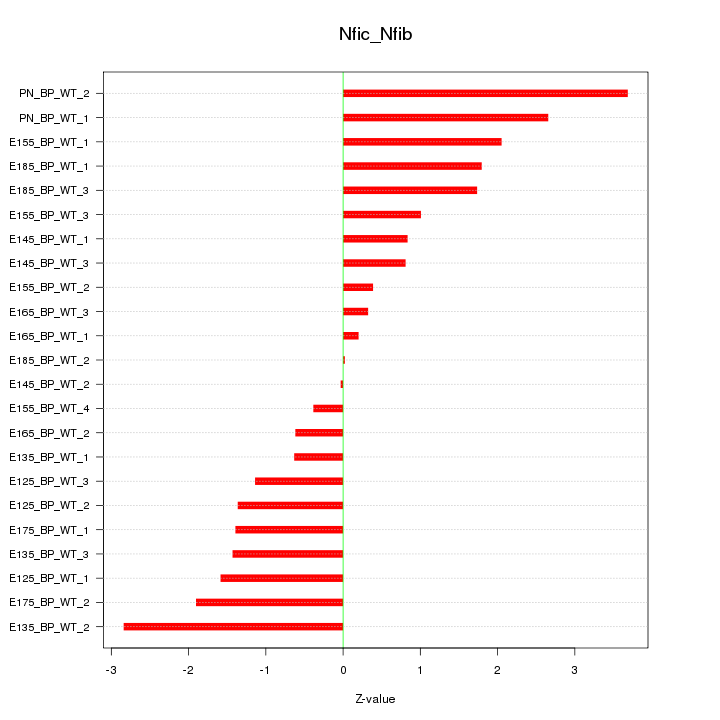
Motif ID: Nfic_Nfib
Z-value: 1.561
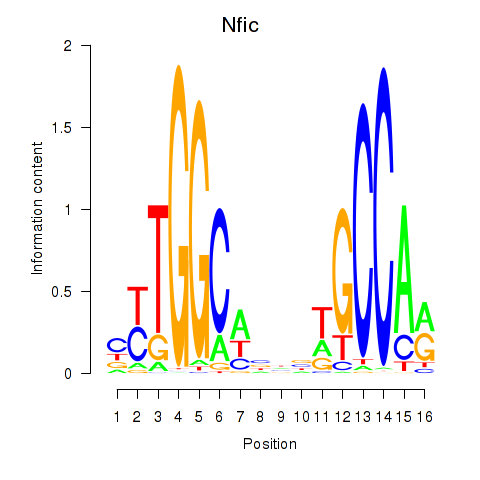
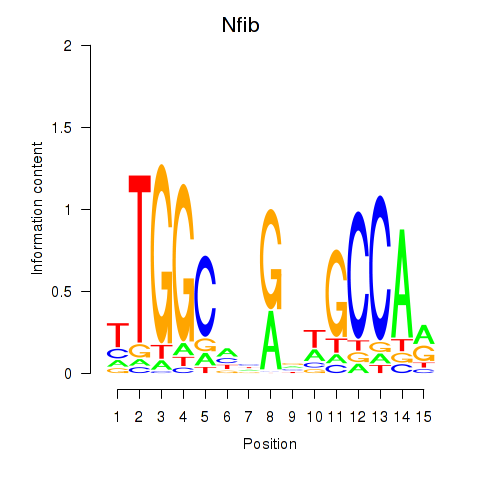
Transcription factors associated with Nfic_Nfib:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfib | ENSMUSG00000008575.11 | Nfib |
| Nfic | ENSMUSG00000055053.11 | Nfic |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfic | mm10_v2_chr10_-_81427114_81427187 | 0.52 | 1.0e-02 | Click! |
| Nfib | mm10_v2_chr4_-_82505274_82505358 | 0.40 | 6.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 24.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 2.3 | 6.8 | GO:0003032 | detection of oxygen(GO:0003032) |
| 1.8 | 5.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.4 | 11.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.4 | 6.9 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 1.2 | 3.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.0 | 6.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 1.0 | 6.0 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.9 | 2.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) |
| 0.9 | 7.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 | 8.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.8 | 2.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 2.3 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.7 | 2.1 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.7 | 2.1 | GO:0072180 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.7 | 2.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.6 | 3.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.6 | 1.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.5 | 3.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.5 | 2.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.5 | 2.1 | GO:0010046 | arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.5 | 2.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.5 | 1.4 | GO:0072071 | positive regulation of glomerular filtration(GO:0003104) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) phenotypic switching(GO:0036166) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.5 | 1.9 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.4 | 1.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 2.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.4 | 1.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 6.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.4 | 1.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.3 | 0.3 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.3 | 1.3 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 2.3 | GO:0051461 | positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 1.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 3.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.3 | 1.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 1.9 | GO:0002317 | plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 0.9 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid catabolic process(GO:0019478) |
| 0.3 | 1.4 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 2.7 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 2.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 0.5 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 2.5 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.2 | 0.9 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 1.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.2 | 0.7 | GO:2000562 | negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 2.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 2.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.2 | 0.5 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 | 0.6 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 1.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 0.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 2.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 | 1.1 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) cellular response to hyperoxia(GO:0071455) |
| 0.2 | 0.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 | 2.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 2.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.5 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 2.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.7 | GO:0036151 | negative regulation of fatty acid beta-oxidation(GO:0031999) phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.9 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 | 3.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 | 4.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 2.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 | 0.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.9 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.6 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.8 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.4 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 1.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.9 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 2.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.3 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 3.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.5 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) proline biosynthetic process(GO:0006561) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.2 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.4 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.3 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 1.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.3 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.5 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 1.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 3.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.5 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.7 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.6 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 1.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.2 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 2.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 2.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.9 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.0 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.5 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.2 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 1.6 | 4.9 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.3 | 4.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.2 | 3.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.1 | 4.5 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.9 | 6.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.6 | 2.5 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.5 | 6.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 0.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 2.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 19.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 0.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 1.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 0.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 2.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 2.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 4.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 5.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 4.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 3.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 14.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 6.2 | GO:0044297 | cell body(GO:0044297) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 2.3 | 6.8 | GO:0005534 | galactose binding(GO:0005534) |
| 1.5 | 8.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.4 | 6.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.4 | 5.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.2 | 4.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 4.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.7 | 2.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 8.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 2.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 1.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) aspartate oxidase activity(GO:0015922) |
| 0.4 | 1.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 3.4 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.4 | 1.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 1.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.3 | 2.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 7.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 1.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.3 | 1.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 0.9 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 1.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 3.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 1.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 6.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 2.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 2.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.9 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 3.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 0.7 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 0.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.5 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.2 | 1.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 5.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 3.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.4 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.1 | 0.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 4.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 2.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 5.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 7.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.3 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 4.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.2 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.3 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:0023029 | peptide antigen-transporting ATPase activity(GO:0015433) MHC class Ib protein binding(GO:0023029) tapasin binding(GO:0046980) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.3 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.5 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |