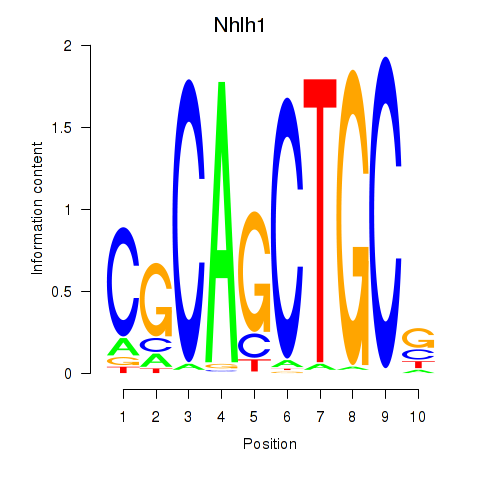
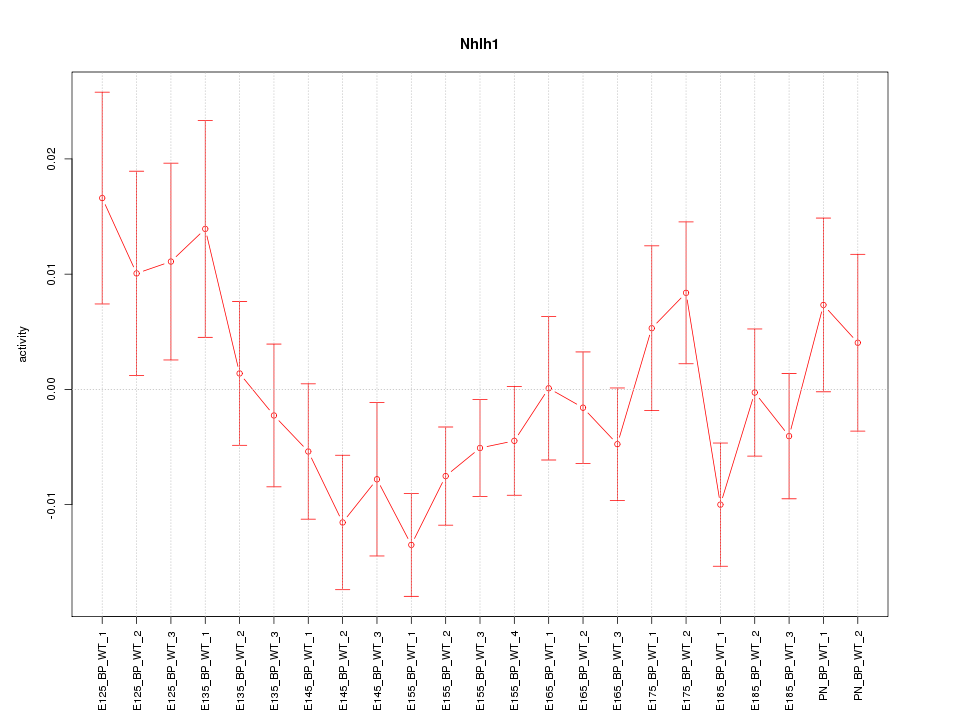
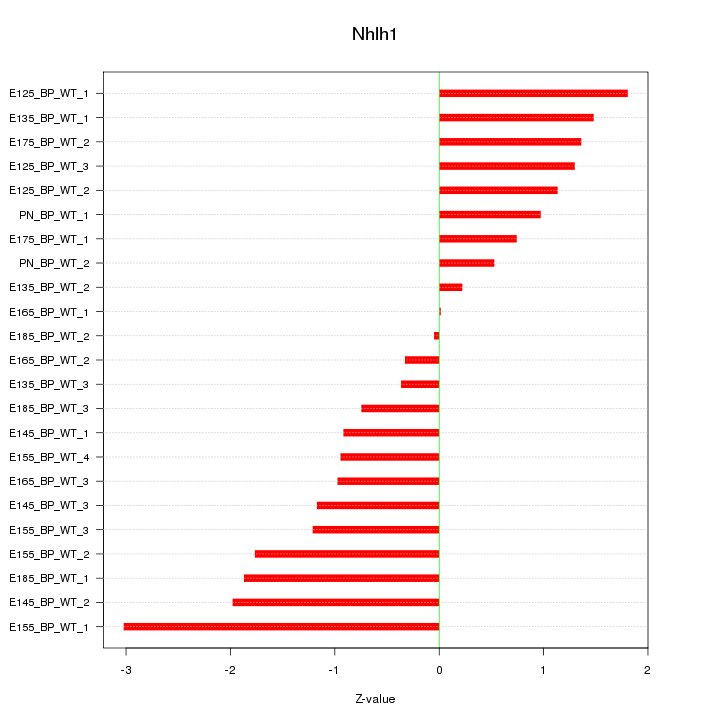
| 0.7 |
2.0 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.6 |
1.9 |
GO:0072076 |
hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.6 |
1.9 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.5 |
2.6 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.5 |
1.4 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.5 |
0.5 |
GO:1900133 |
renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) |
| 0.4 |
1.8 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 |
1.8 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.4 |
1.3 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.4 |
1.3 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 |
1.6 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.4 |
1.2 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.4 |
1.5 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.4 |
2.6 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.3 |
1.0 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.3 |
0.7 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.3 |
1.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 |
2.6 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 |
1.9 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.3 |
2.2 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 |
1.2 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.3 |
0.9 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.3 |
2.7 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.3 |
1.5 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 |
0.6 |
GO:0010512 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.3 |
1.2 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.3 |
1.5 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.3 |
0.9 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.3 |
0.6 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 |
0.9 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.3 |
0.9 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 |
1.4 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.3 |
2.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.3 |
1.7 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 |
0.8 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.3 |
0.3 |
GO:0034135 |
regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.3 |
0.8 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 |
0.8 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 |
1.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
0.7 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 |
1.7 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.2 |
0.7 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 |
1.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 |
0.7 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.2 |
2.7 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 |
0.6 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 |
0.4 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.2 |
0.6 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.2 |
2.5 |
GO:0001977 |
renal system process involved in regulation of blood volume(GO:0001977) |
| 0.2 |
0.4 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.2 |
0.6 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 |
2.0 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.2 |
1.4 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 |
0.2 |
GO:0048819 |
regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) |
| 0.2 |
1.3 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 |
0.6 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 |
0.7 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 |
1.3 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 |
0.5 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 |
1.8 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.2 |
0.5 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.2 |
1.2 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
0.2 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.2 |
0.7 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 |
0.5 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 |
0.5 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.2 |
0.5 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.2 |
0.3 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.2 |
1.5 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 |
0.5 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.2 |
0.8 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
1.4 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) |
| 0.2 |
0.2 |
GO:0061054 |
dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 |
0.6 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
1.8 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.4 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 |
0.7 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.7 |
GO:0001865 |
NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 |
0.3 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 |
3.2 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.1 |
0.7 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.8 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.1 |
0.3 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 |
0.3 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 |
1.5 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
1.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.9 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 |
0.4 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.1 |
1.4 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.5 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.4 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
0.2 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.1 |
0.4 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
0.9 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
0.7 |
GO:0071600 |
otic vesicle morphogenesis(GO:0071600) |
| 0.1 |
0.5 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 |
0.6 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
0.4 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 |
1.2 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
2.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.3 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.1 |
0.8 |
GO:0021747 |
cochlear nucleus development(GO:0021747) |
| 0.1 |
0.3 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.2 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
1.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.9 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.1 |
0.8 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.5 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
2.2 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.1 |
0.4 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.1 |
0.5 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
1.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.4 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.8 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.6 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.1 |
0.4 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.1 |
0.4 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.3 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.2 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.1 |
0.5 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.3 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.3 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 |
0.6 |
GO:0051572 |
negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 |
0.6 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.4 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 |
0.3 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.1 |
0.4 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.1 |
0.9 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
1.1 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.3 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
0.3 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
0.8 |
GO:0002523 |
leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 |
0.2 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 |
0.5 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
1.8 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
0.6 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.4 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.2 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.2 |
GO:0032079 |
positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 |
0.2 |
GO:0071354 |
response to interleukin-6(GO:0070741) cellular response to interleukin-6(GO:0071354) |
| 0.1 |
0.7 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 |
0.5 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.5 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.3 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.1 |
0.3 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 |
1.6 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
0.4 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.1 |
0.4 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.5 |
GO:0051770 |
positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 |
0.2 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
2.2 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
0.2 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
1.2 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.1 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 |
0.6 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) |
| 0.1 |
0.3 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.3 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.1 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.4 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
2.2 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.3 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.1 |
0.9 |
GO:0007213 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.3 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 |
0.5 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.5 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.1 |
0.6 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.1 |
0.3 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.2 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.4 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
0.5 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
1.5 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.1 |
0.3 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 |
0.1 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.2 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.3 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.5 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.3 |
GO:0097680 |
double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 |
0.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 |
0.6 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.6 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
0.2 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 |
0.5 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.2 |
GO:0006661 |
phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 |
0.8 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
0.6 |
GO:2000254 |
regulation of male germ cell proliferation(GO:2000254) |
| 0.1 |
0.2 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
1.0 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.1 |
0.2 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 |
1.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.2 |
GO:0006507 |
GPI anchor release(GO:0006507) |
| 0.1 |
0.2 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.3 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.6 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.2 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 |
0.2 |
GO:0015744 |
succinate transport(GO:0015744) |
| 0.1 |
0.3 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
0.4 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.4 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
1.2 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.1 |
1.3 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.6 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.3 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
2.1 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.8 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.4 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:1904683 |
regulation of metalloendopeptidase activity(GO:1904683) negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 |
0.5 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.0 |
0.6 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.4 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 |
0.3 |
GO:1904294 |
positive regulation of ERAD pathway(GO:1904294) |
| 0.0 |
0.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.0 |
GO:0002586 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 |
0.3 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.7 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.9 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.2 |
GO:1903223 |
positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 |
0.1 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.0 |
0.4 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.3 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
1.6 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.2 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.5 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.2 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.4 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.5 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0097411 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 |
0.5 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.4 |
GO:0021670 |
lateral ventricle development(GO:0021670) |
| 0.0 |
0.8 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.3 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.6 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.2 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 |
0.2 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.4 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.4 |
GO:0021684 |
cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.1 |
GO:0045144 |
female meiosis sister chromatid cohesion(GO:0007066) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 |
0.2 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.2 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
1.1 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.3 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.0 |
0.4 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.2 |
GO:0030423 |
targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 |
0.7 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.2 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.5 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.3 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 |
0.4 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.8 |
GO:0070098 |
chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 |
0.2 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
0.2 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.7 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.2 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.1 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.1 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.4 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
1.0 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) |
| 0.0 |
0.2 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.1 |
GO:0045616 |
regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 |
0.7 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.1 |
GO:0003085 |
negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 |
0.4 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.9 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.3 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.0 |
0.1 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.2 |
GO:0006586 |
tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.0 |
0.2 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 |
0.1 |
GO:0050996 |
positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 |
0.7 |
GO:0044030 |
regulation of DNA methylation(GO:0044030) |
| 0.0 |
0.3 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 |
0.2 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
1.5 |
GO:0051304 |
chromosome separation(GO:0051304) |
| 0.0 |
0.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.0 |
GO:0006591 |
ornithine metabolic process(GO:0006591) |
| 0.0 |
0.2 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.0 |
0.4 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.2 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.0 |
0.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.4 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.6 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
0.3 |
GO:1900048 |
positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 |
0.4 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.3 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.1 |
GO:0002756 |
MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 |
0.0 |
GO:0072053 |
establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 |
0.7 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.4 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.0 |
0.3 |
GO:0033866 |
coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 |
1.4 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.1 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 |
0.1 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 |
0.4 |
GO:0031111 |
negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 |
0.2 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.2 |
GO:0045620 |
negative regulation of lymphocyte differentiation(GO:0045620) |
| 0.0 |
0.8 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.0 |
0.3 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.2 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.4 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 |
0.2 |
GO:0006534 |
cysteine metabolic process(GO:0006534) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.1 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.0 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.4 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.1 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.2 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.1 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.6 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.2 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.4 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.7 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.1 |
GO:0032695 |
negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 |
0.2 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.0 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 |
0.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
1.6 |
GO:0050821 |
protein stabilization(GO:0050821) |
| 0.0 |
0.2 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.0 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.0 |
0.1 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.1 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.3 |
GO:0098868 |
endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.0 |
0.3 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.2 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.3 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.0 |
0.2 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.4 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.2 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.1 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.1 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.4 |
GO:0055007 |
cardiac muscle cell differentiation(GO:0055007) |
| 0.0 |
0.1 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0051155 |
positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 |
0.3 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.1 |
GO:0051569 |
regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 |
0.1 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 |
0.2 |
GO:1902042 |
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 |
0.2 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.0 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 |
0.2 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
1.2 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.0 |
0.1 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 |
0.3 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.0 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.1 |
GO:0018377 |
N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.2 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
1.1 |
GO:0016331 |
morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.1 |
GO:0046856 |
phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.0 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.1 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.1 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.4 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.0 |
0.1 |
GO:0060481 |
bronchus development(GO:0060433) lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 |
0.1 |
GO:0071897 |
DNA biosynthetic process(GO:0071897) |
| 0.0 |
0.1 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.1 |
GO:0001706 |
endoderm formation(GO:0001706) |