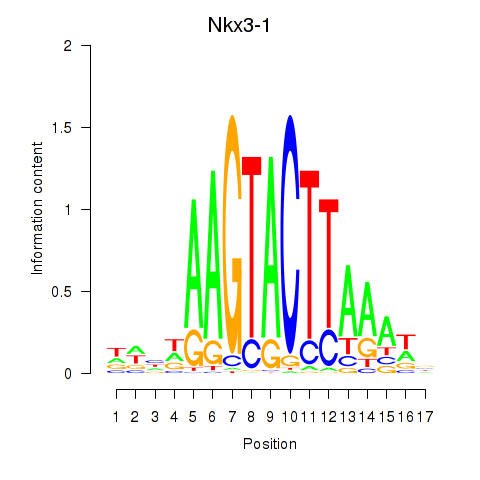
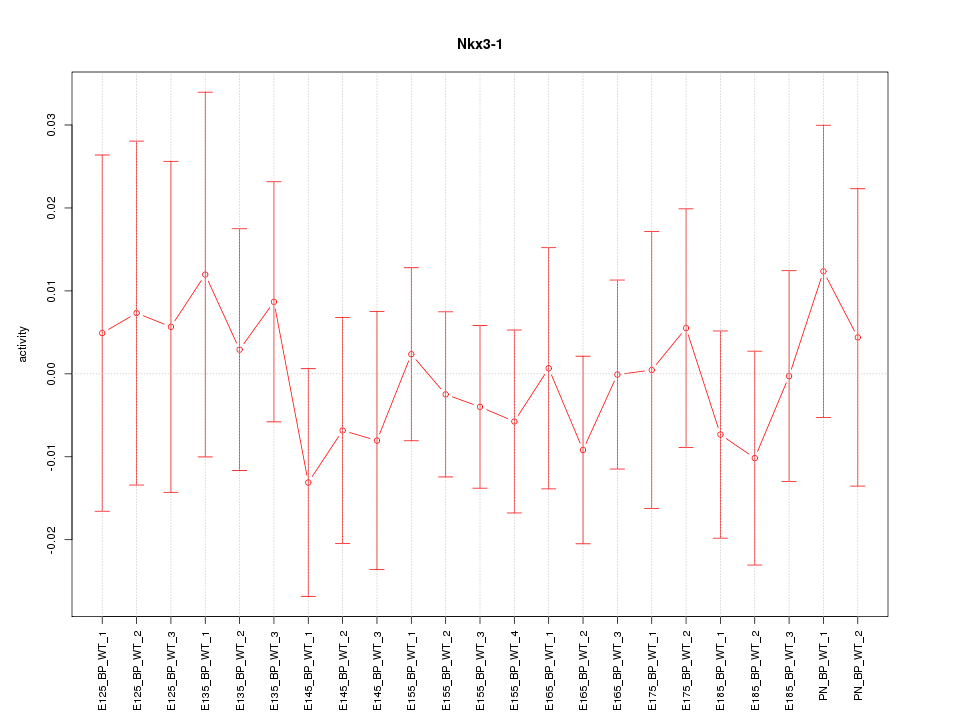
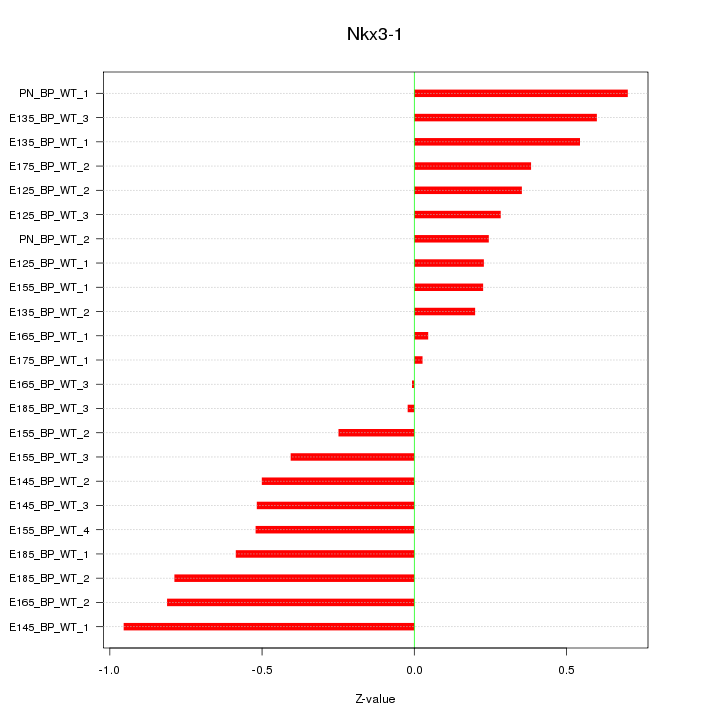
| 0.3 |
0.8 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.2 |
0.9 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 |
0.5 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.6 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.1 |
0.4 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.9 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.4 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 |
0.6 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.3 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.1 |
0.5 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.3 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
0.3 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.4 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.3 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 |
0.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
1.0 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.5 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.9 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.2 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 |
0.4 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
0.4 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 |
0.2 |
GO:0002666 |
positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 |
0.1 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.0 |
0.2 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) amylase secretion(GO:0036394) |
| 0.0 |
0.3 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.1 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.0 |
0.1 |
GO:2000407 |
positive regulation of necrotic cell death(GO:0010940) regulation of T cell extravasation(GO:2000407) |
| 0.0 |
0.1 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 |
0.3 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.1 |
GO:1901162 |
tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 |
0.2 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.5 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.4 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.3 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.3 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.1 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.9 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.2 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.0 |
0.1 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |