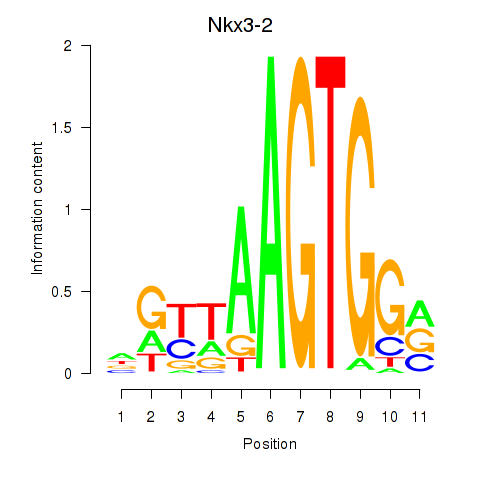
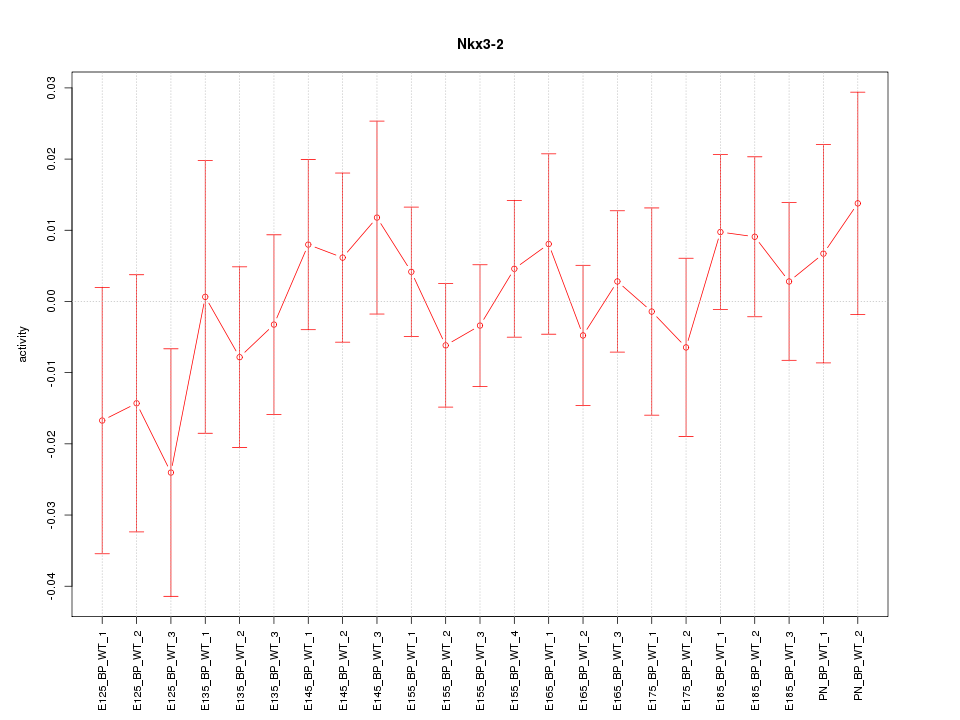
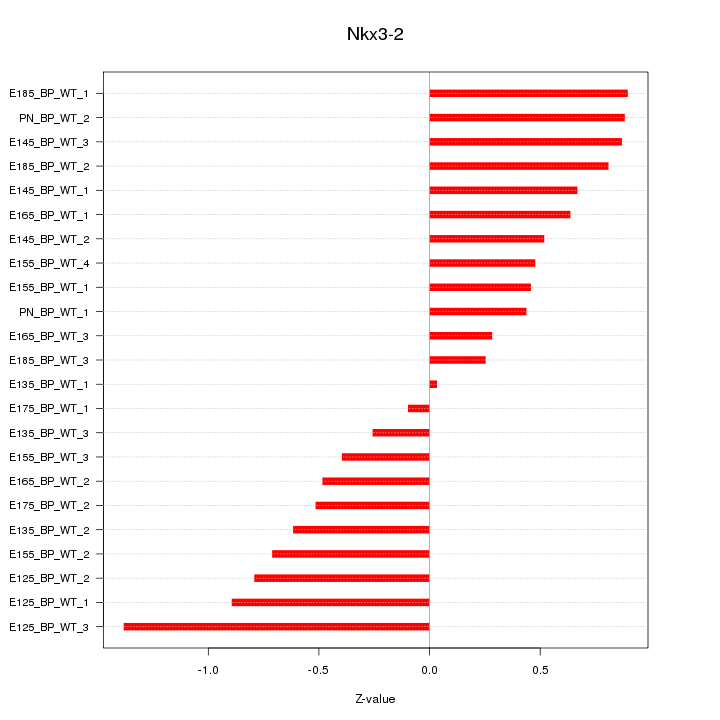
| 0.4 |
1.9 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.4 |
1.1 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 |
1.1 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 |
1.5 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.3 |
1.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.3 |
0.8 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 |
1.0 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 |
0.7 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 |
1.4 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.2 |
0.6 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 |
1.3 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 |
1.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
0.5 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 |
0.8 |
GO:0044838 |
cell quiescence(GO:0044838) |
| 0.2 |
2.2 |
GO:0071371 |
cellular response to electrical stimulus(GO:0071257) cellular response to gonadotropin stimulus(GO:0071371) |
| 0.2 |
0.7 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 |
1.5 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 |
1.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 |
3.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
1.3 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 |
0.3 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.1 |
0.4 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.7 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 |
1.1 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.2 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.5 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 |
0.7 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
0.5 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.3 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.1 |
0.2 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.3 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.2 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 |
0.5 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
0.9 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.1 |
0.9 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.4 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
2.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.4 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 |
1.5 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.6 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 |
0.4 |
GO:0070813 |
hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 |
1.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.2 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.8 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.3 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.4 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.2 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.2 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 |
0.6 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.2 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.4 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.4 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.2 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 |
0.5 |
GO:0031272 |
regulation of pseudopodium assembly(GO:0031272) |
| 0.0 |
0.7 |
GO:0046321 |
positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
1.2 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.4 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.3 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
0.4 |
GO:0002237 |
response to molecule of bacterial origin(GO:0002237) response to lipopolysaccharide(GO:0032496) |
| 0.0 |
0.6 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.6 |
GO:0001963 |
synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 |
0.3 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
1.7 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
1.5 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.6 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.0 |
0.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
1.0 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 |
0.3 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.8 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.8 |
GO:0006635 |
fatty acid beta-oxidation(GO:0006635) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.3 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.3 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.0 |
GO:0002074 |
extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |