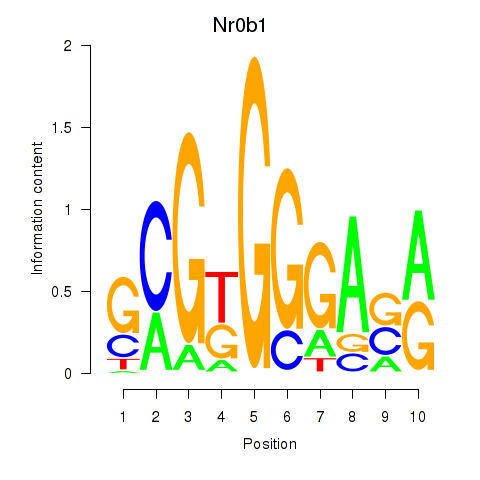
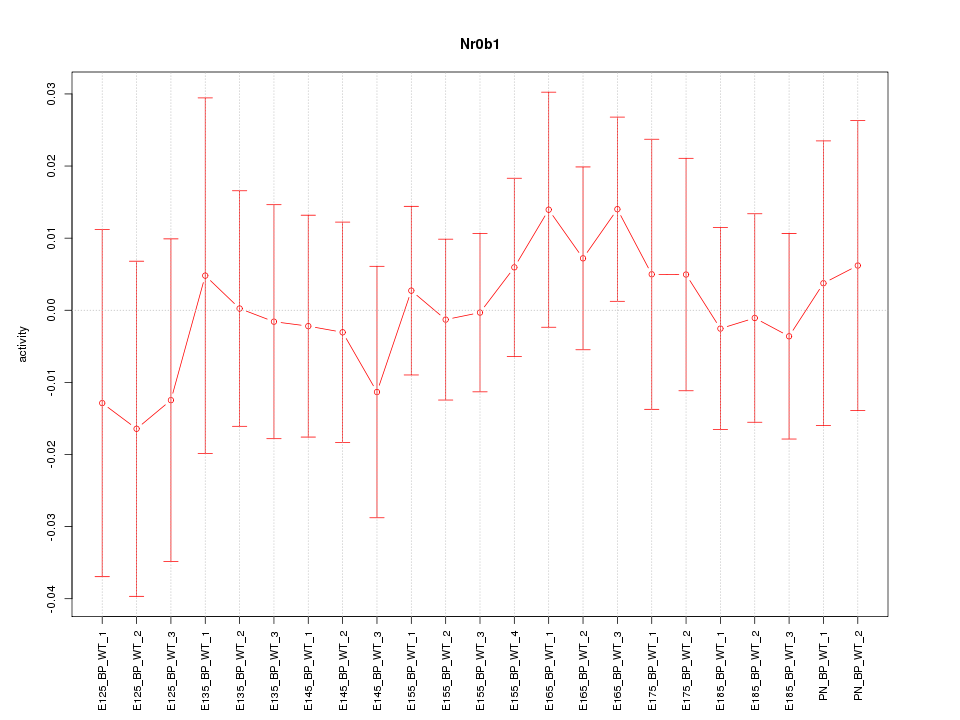
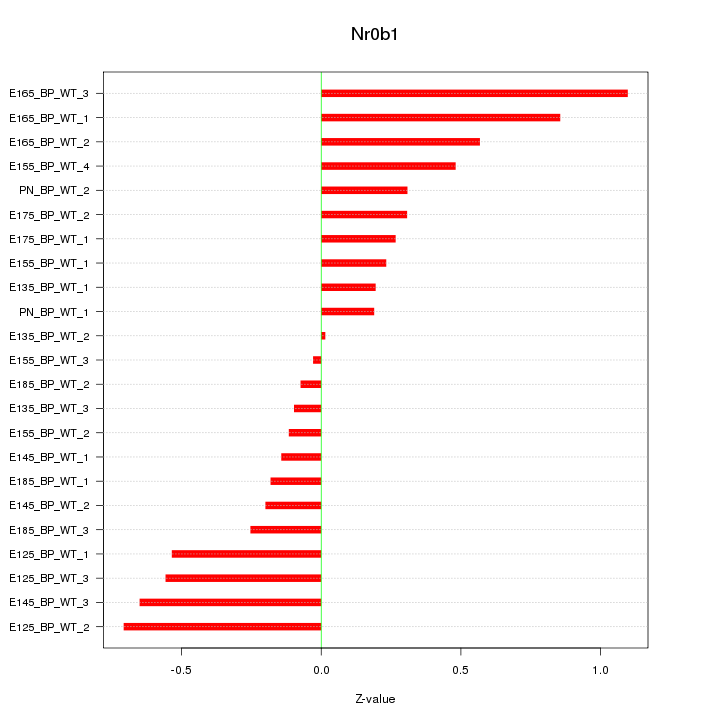
| 0.6 |
1.7 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.1 |
0.5 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
1.5 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.4 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
0.4 |
GO:0048007 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 |
0.3 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 |
0.6 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
1.5 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.2 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.2 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.1 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) |
| 0.0 |
0.2 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.0 |
0.1 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 |
0.3 |
GO:0042996 |
regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.6 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.4 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.6 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.4 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.4 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.1 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.0 |
0.1 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 |
0.2 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.6 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.6 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.3 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.0 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.4 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |