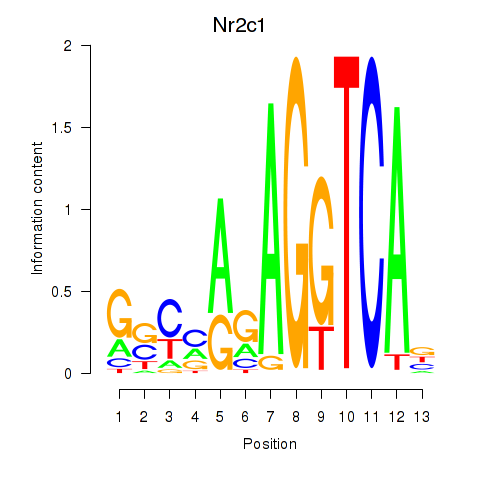
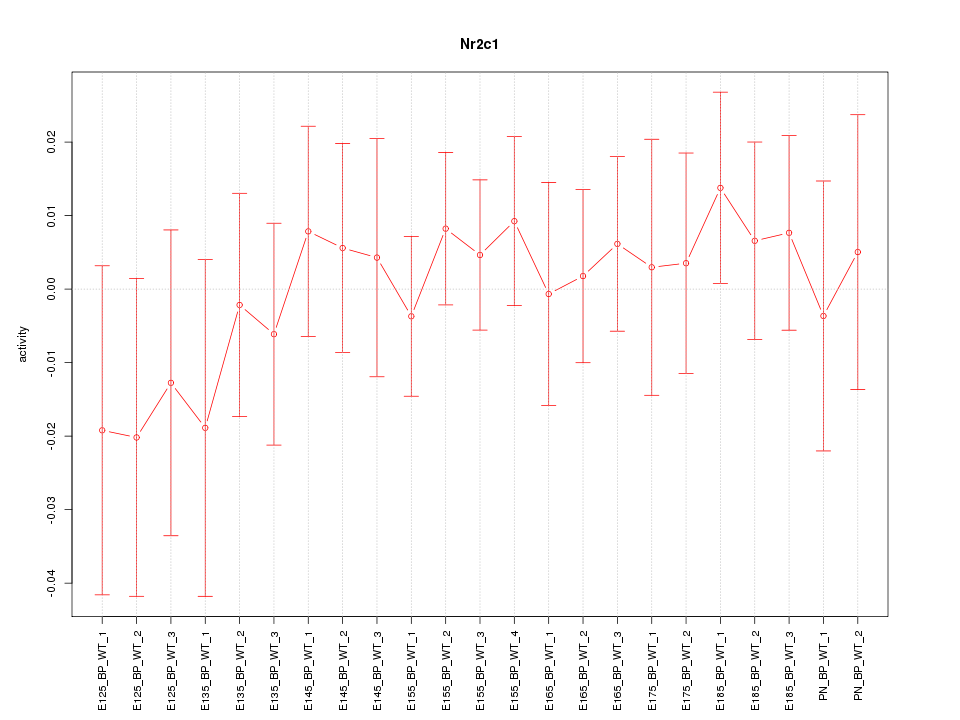
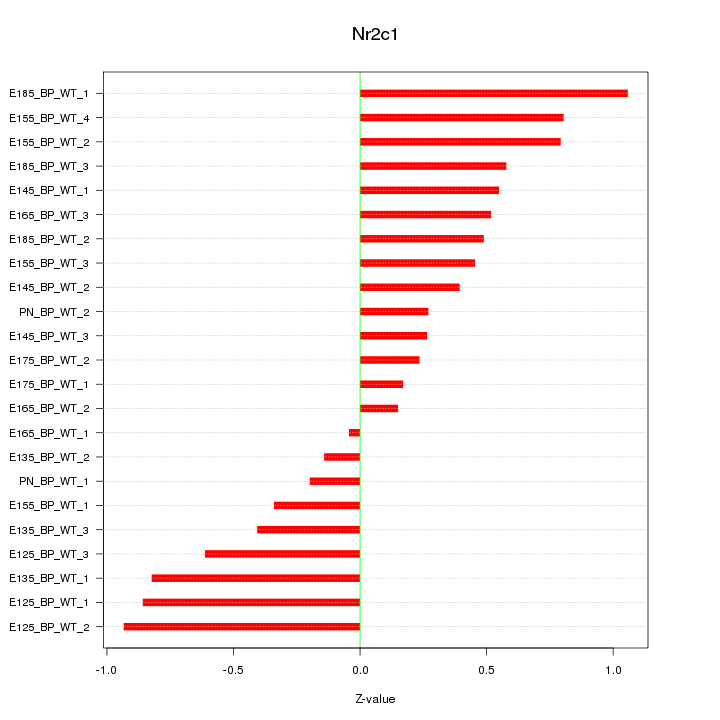
| 0.5 |
1.4 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.4 |
0.7 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 |
1.6 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
0.8 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
0.7 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.4 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 |
0.4 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.7 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
1.0 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.5 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 |
1.9 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.3 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.3 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.3 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 |
0.5 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
0.2 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 |
0.3 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 |
1.3 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.1 |
0.3 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.2 |
GO:0070973 |
COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.8 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.4 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.2 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.2 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 |
0.9 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.4 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.4 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.3 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.1 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
1.3 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.8 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.1 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.0 |
1.4 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.1 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.5 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.3 |
GO:0031069 |
hair follicle morphogenesis(GO:0031069) |
| 0.0 |
0.6 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.5 |
GO:0097006 |
regulation of plasma lipoprotein particle levels(GO:0097006) |