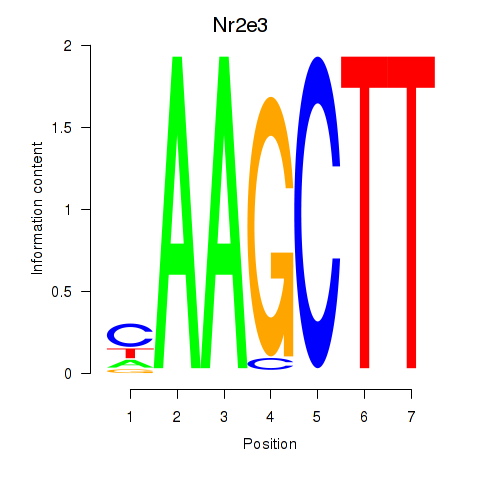
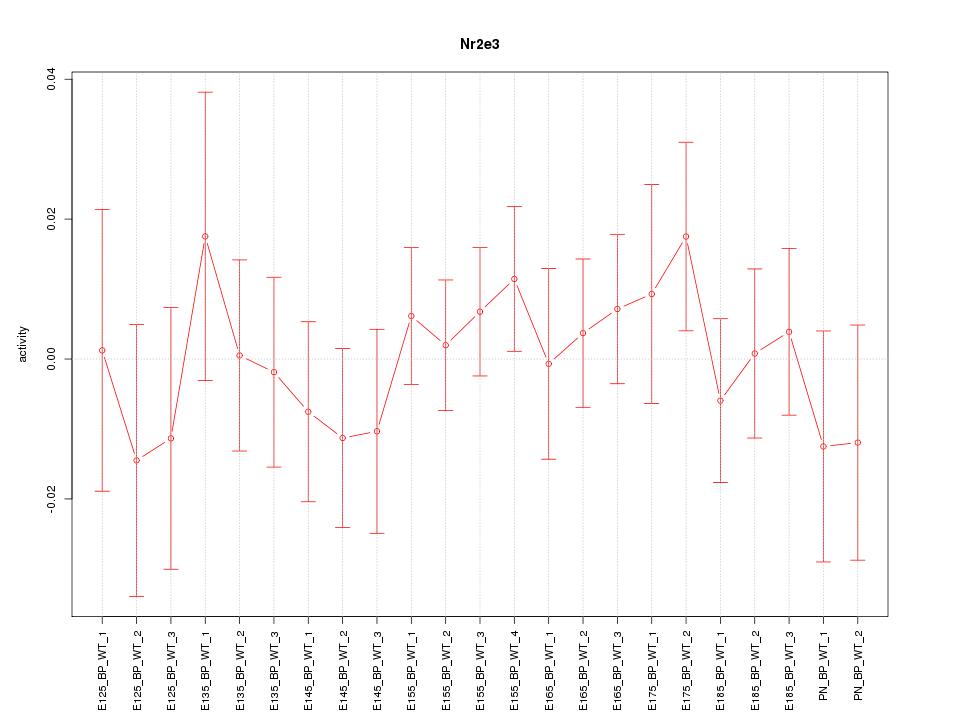
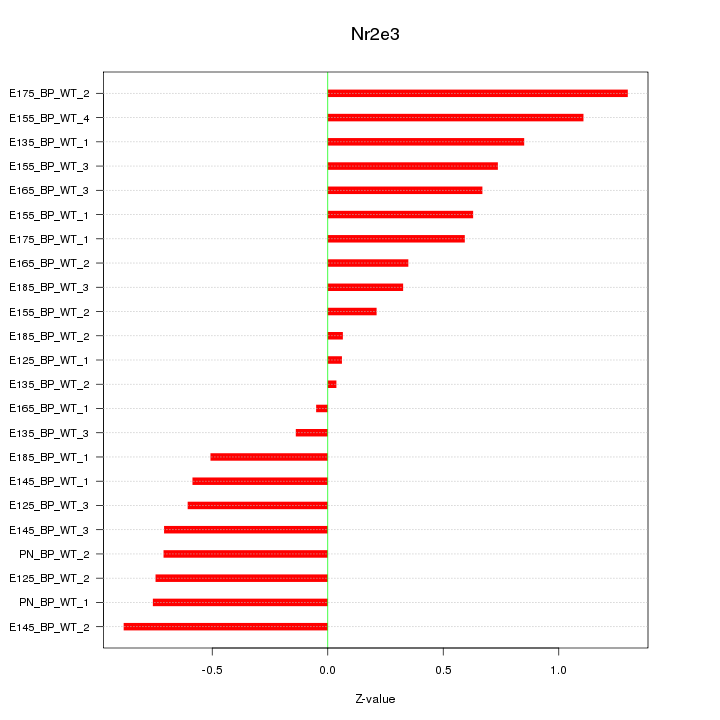
| 0.7 |
2.2 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.2 |
1.1 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 |
0.6 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 |
0.6 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.7 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.9 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.6 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.9 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.3 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 |
0.5 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.1 |
2.2 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
0.3 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
2.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.7 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.7 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 |
0.2 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.6 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.4 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.4 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.5 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.4 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.3 |
GO:0036508 |
protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 |
0.5 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.6 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.5 |
GO:0051791 |
medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 |
0.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
1.1 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.2 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.8 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.4 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 |
0.2 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.1 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 |
0.2 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.1 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
0.1 |
GO:0042713 |
operant conditioning(GO:0035106) sperm ejaculation(GO:0042713) |
| 0.0 |
0.5 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.1 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.4 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.4 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.0 |
0.1 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.0 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 |
0.8 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.5 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.0 |
0.2 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.3 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.2 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.0 |
0.1 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
1.0 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.6 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |