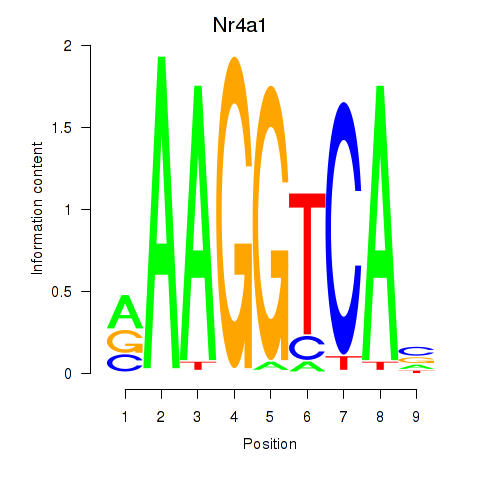
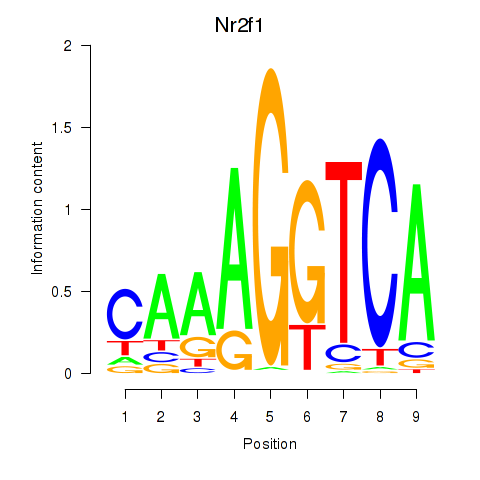
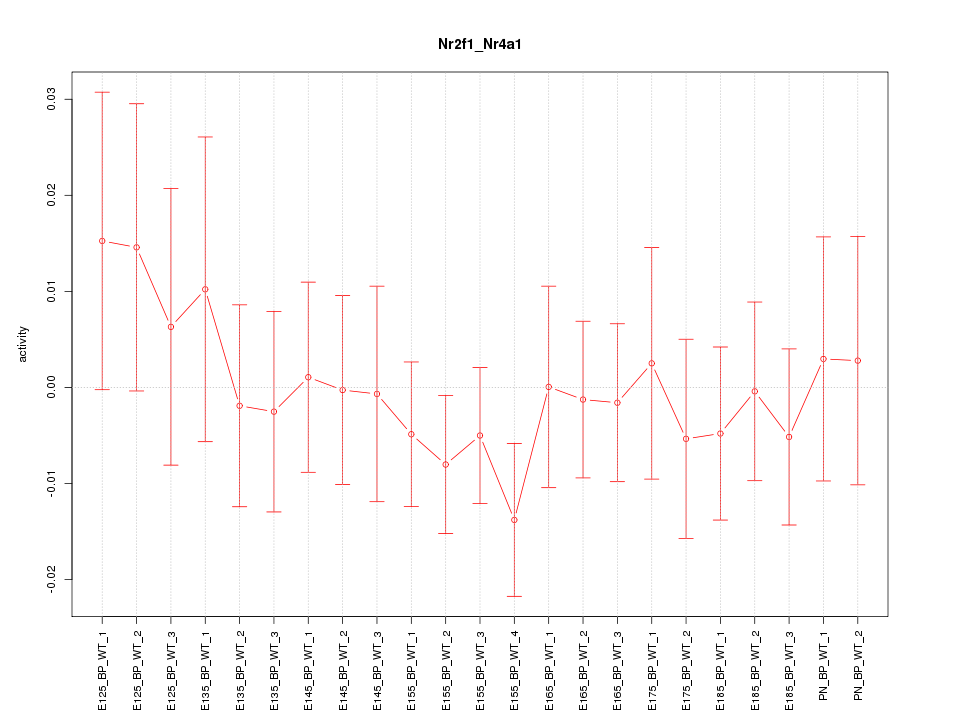
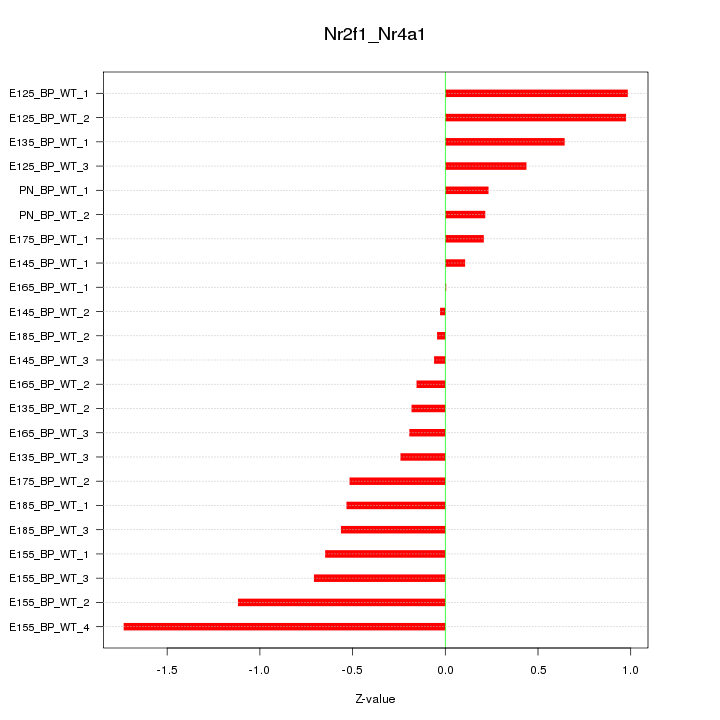
| 0.7 |
2.0 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.7 |
2.0 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.4 |
1.7 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.4 |
0.8 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.4 |
1.1 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.4 |
1.4 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.3 |
1.0 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.2 |
0.9 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.2 |
0.7 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.2 |
0.7 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.2 |
1.4 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.2 |
0.2 |
GO:0097491 |
trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.2 |
1.0 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 |
1.0 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 |
1.2 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 |
0.6 |
GO:0060455 |
regulation of gastric acid secretion(GO:0060453) negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 |
0.4 |
GO:1903275 |
positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.2 |
0.5 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 |
1.0 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 |
0.5 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 |
0.8 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 |
0.8 |
GO:0090234 |
cellular response to testosterone stimulus(GO:0071394) regulation of kinetochore assembly(GO:0090234) |
| 0.2 |
0.5 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 |
2.9 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 |
0.8 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
0.5 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
0.7 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
1.3 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 |
0.5 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 |
0.4 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) hematopoietic stem cell migration(GO:0035701) regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
0.3 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.5 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.4 |
GO:0006145 |
purine nucleobase catabolic process(GO:0006145) |
| 0.1 |
1.5 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
0.3 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.1 |
0.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.3 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.2 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.3 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 0.1 |
0.5 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.1 |
GO:0034140 |
negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 |
0.3 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.6 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.8 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.3 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 |
0.3 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 |
0.5 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
0.8 |
GO:0035584 |
calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 |
0.8 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
0.7 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.2 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 |
0.2 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
0.3 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
0.2 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 |
3.3 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
0.4 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
0.9 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.2 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
0.2 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.2 |
GO:0061198 |
fungiform papilla formation(GO:0061198) |
| 0.1 |
0.2 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.1 |
GO:1901295 |
regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 |
0.1 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.2 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
0.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.2 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 |
0.4 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 |
0.4 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
0.3 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 |
0.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
0.4 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.3 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.4 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.6 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 |
0.6 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.1 |
0.5 |
GO:0043586 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) tongue development(GO:0043586) |
| 0.1 |
1.8 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.1 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) negative regulation of female gonad development(GO:2000195) |
| 0.0 |
0.2 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 |
0.3 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
0.1 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 |
0.4 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.3 |
GO:0097066 |
response to thyroid hormone(GO:0097066) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.4 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.5 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.1 |
GO:0010579 |
regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) regulation of corticosterone secretion(GO:2000852) |
| 0.0 |
1.1 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.1 |
GO:1902336 |
coronary vein morphogenesis(GO:0003169) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 |
0.9 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
1.2 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.4 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.9 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.1 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.0 |
0.2 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 |
0.2 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) macropinocytosis(GO:0044351) |
| 0.0 |
0.3 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 |
0.1 |
GO:0046078 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.3 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.1 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 |
0.5 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.7 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.9 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.1 |
GO:1902947 |
regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.3 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 |
0.3 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.4 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 |
0.1 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.0 |
0.5 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.4 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.1 |
GO:0038030 |
non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 |
0.9 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.2 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 |
0.5 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
1.0 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.1 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 |
0.2 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.1 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 |
0.3 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
1.0 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.2 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 |
2.0 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.1 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.2 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 |
0.3 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0044830 |
modulation by host of viral RNA genome replication(GO:0044830) |
| 0.0 |
0.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.1 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.1 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.2 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.0 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
0.1 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 |
0.0 |
GO:0003160 |
endocardium morphogenesis(GO:0003160) |
| 0.0 |
0.3 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 |
0.4 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.4 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.1 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 |
0.1 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.0 |
0.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.1 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.6 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.1 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.0 |
0.2 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.0 |
0.1 |
GO:1902277 |
negative regulation of growth hormone secretion(GO:0060125) negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 |
0.6 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.1 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.1 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
0.6 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.2 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.2 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
2.3 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.1 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.5 |
GO:0031111 |
negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 |
1.2 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 |
0.2 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.1 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.1 |
GO:0045346 |
regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 |
0.2 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
0.0 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) |
| 0.0 |
0.3 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.3 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.3 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.3 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.1 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.3 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.0 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.1 |
GO:0042407 |
inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.0 |
0.0 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 |
0.0 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.0 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.4 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.3 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.0 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 |
0.3 |
GO:0042026 |
protein refolding(GO:0042026) |
| 0.0 |
0.0 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 |
0.2 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.1 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.3 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
1.0 |
GO:0000086 |
G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 |
0.3 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.2 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.2 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.2 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.0 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.3 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |