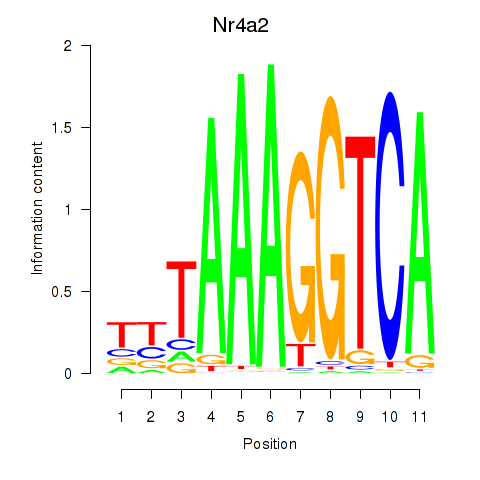
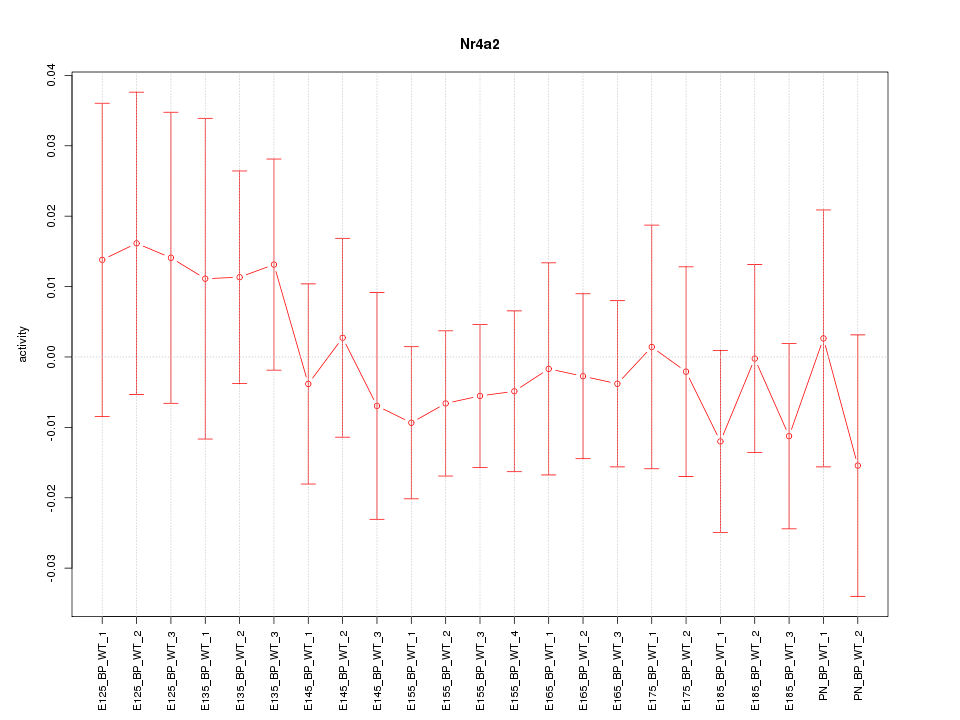
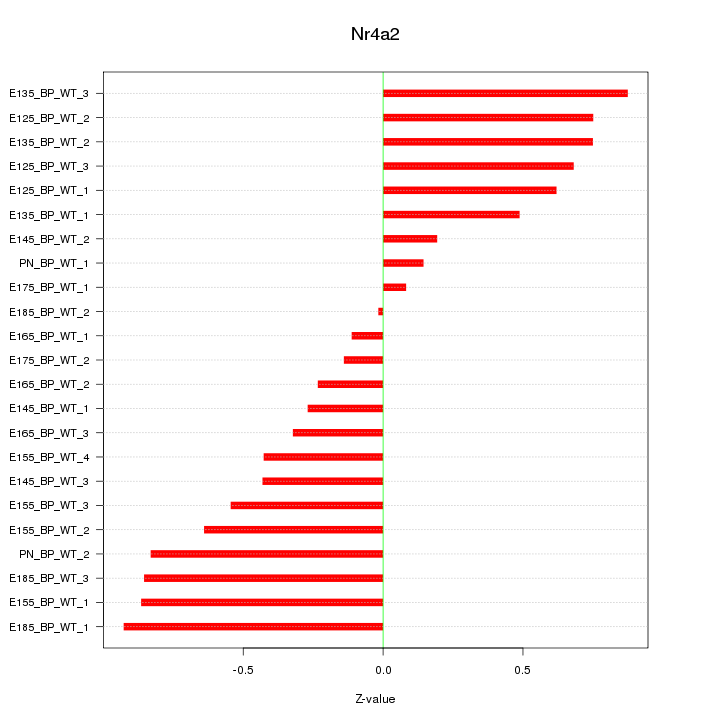
| 0.3 |
0.9 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.2 |
0.6 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
0.5 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
2.0 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
0.4 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) hematopoietic stem cell migration(GO:0035701) regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
0.4 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
0.3 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.1 |
0.3 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
0.3 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 |
0.3 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
2.0 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
1.1 |
GO:0048875 |
chemical homeostasis within a tissue(GO:0048875) |
| 0.1 |
0.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.6 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
0.6 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
2.1 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
0.5 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.5 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.2 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
4.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.3 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
1.2 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
1.4 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.5 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.2 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.2 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.2 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.2 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.4 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.2 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 |
0.1 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.0 |
GO:0032829 |
regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.0 |
0.1 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.0 |
0.6 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 |
0.2 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.1 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.0 |
0.2 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.4 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.5 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.0 |
GO:0070973 |
COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.5 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.9 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |