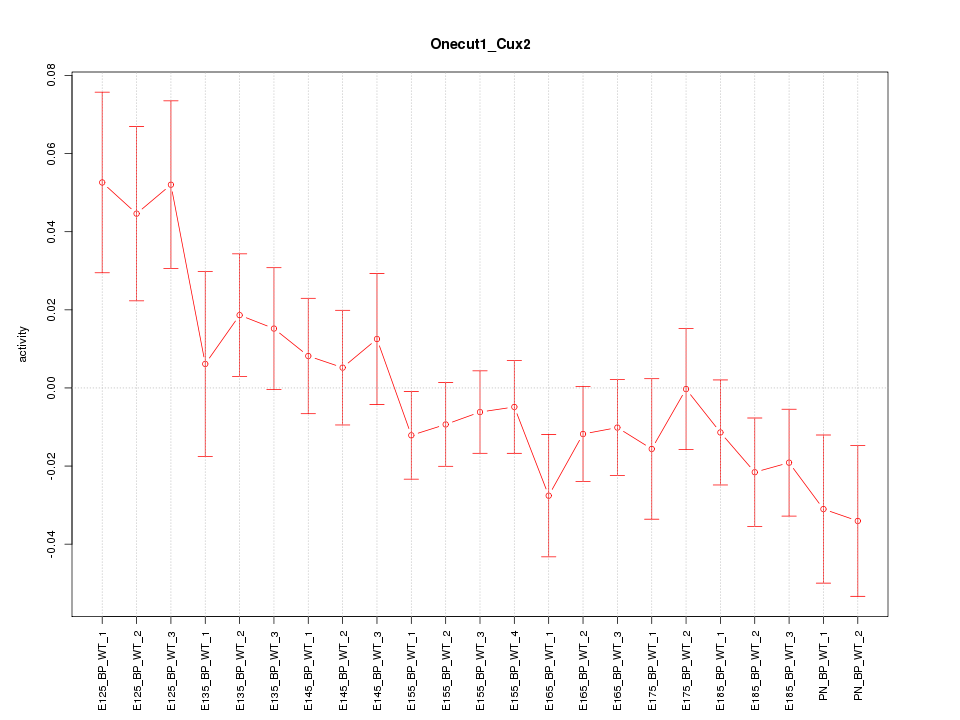
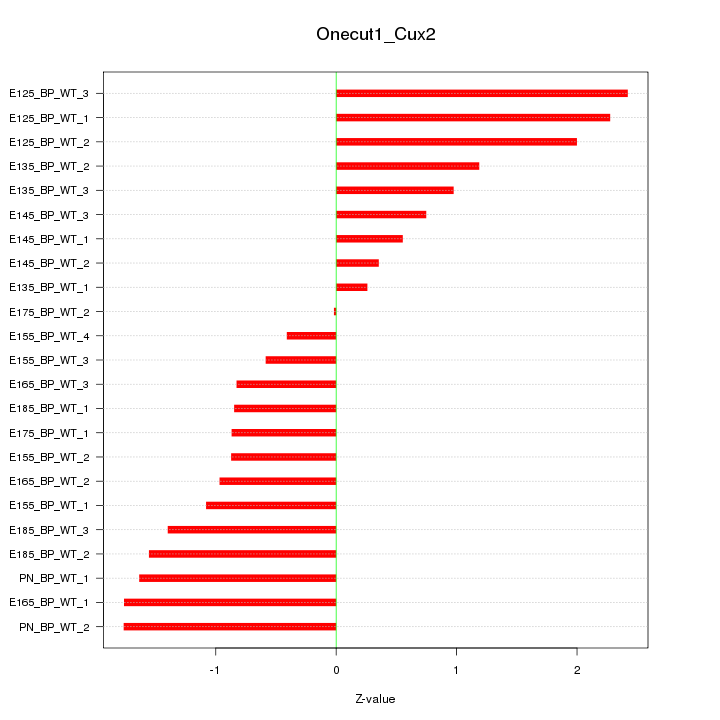
| 1.0 |
7.3 |
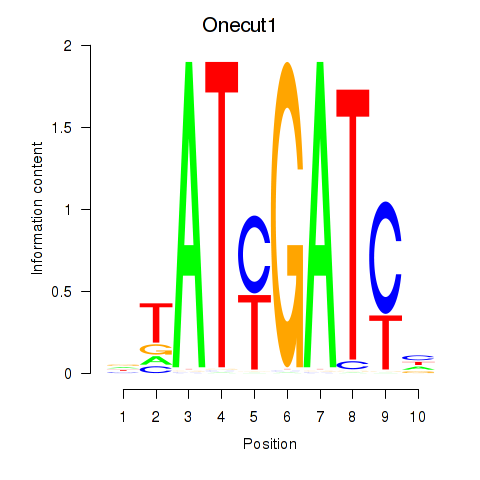
GO:0015016 |
[heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.6 |
3.4 |
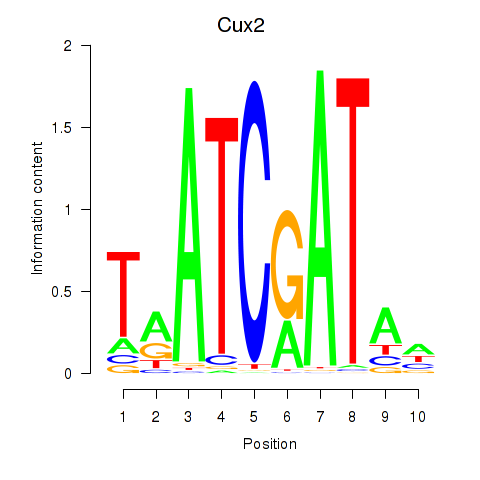
GO:0005042 |
netrin receptor activity(GO:0005042) |
| 0.5 |
12.0 |
GO:0043274 |
phospholipase binding(GO:0043274) |
| 0.5 |
2.3 |
GO:0005052 |
peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 |
1.5 |
GO:0008142 |
oxysterol binding(GO:0008142) |
| 0.4 |
2.5 |
GO:0031802 |
type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 |
0.8 |
GO:0004105 |
choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 |
9.1 |
GO:0001102 |
RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 |
1.7 |
GO:0035925 |
mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 |
0.6 |
GO:0070878 |
primary miRNA binding(GO:0070878) |
| 0.1 |
25.8 |
GO:0001227 |
transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 |
0.3 |
GO:0034739 |
histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 |
9.2 |
GO:0004860 |
protein kinase inhibitor activity(GO:0004860) |
| 0.1 |
3.4 |
GO:0032266 |
phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 |
3.3 |
GO:0035496 |
UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.1 |
0.6 |
GO:0004889 |
acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 |
0.8 |
GO:0051864 |
histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 |
4.0 |
GO:0004222 |
metalloendopeptidase activity(GO:0004222) |
| 0.0 |
0.3 |
GO:0004013 |
adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 |
1.3 |
GO:0034945 |
dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 |
2.3 |
GO:0000979 |
RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 |
0.7 |
GO:0016646 |
oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 |
0.3 |
GO:1990226 |
histone methyltransferase binding(GO:1990226) |
| 0.0 |
1.4 |
GO:0070491 |
repressing transcription factor binding(GO:0070491) |
| 0.0 |
0.2 |
GO:0015375 |
glycine:sodium symporter activity(GO:0015375) |
| 0.0 |
2.1 |
GO:0048306 |
calcium-dependent protein binding(GO:0048306) |
| 0.0 |
0.2 |
GO:0005451 |
monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 |
1.5 |
GO:0031593 |
polyubiquitin binding(GO:0031593) |
| 0.0 |
1.5 |
GO:0043774 |
UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 |
1.0 |
GO:0046875 |
ephrin receptor binding(GO:0046875) |
| 0.0 |
0.5 |
GO:0030506 |
ankyrin binding(GO:0030506) |
| 0.0 |
2.8 |
GO:0003714 |
transcription corepressor activity(GO:0003714) |
| 0.0 |
0.2 |
GO:0005243 |
gap junction channel activity(GO:0005243) |
| 0.0 |
0.3 |
GO:0017091 |
AU-rich element binding(GO:0017091) |
| 0.0 |
15.9 |
GO:0008270 |
zinc ion binding(GO:0008270) |
| 0.0 |
0.9 |
GO:0003725 |
double-stranded RNA binding(GO:0003725) |
| 0.0 |
0.4 |
GO:0050694 |
galactose 3-O-sulfotransferase activity(GO:0050694) |