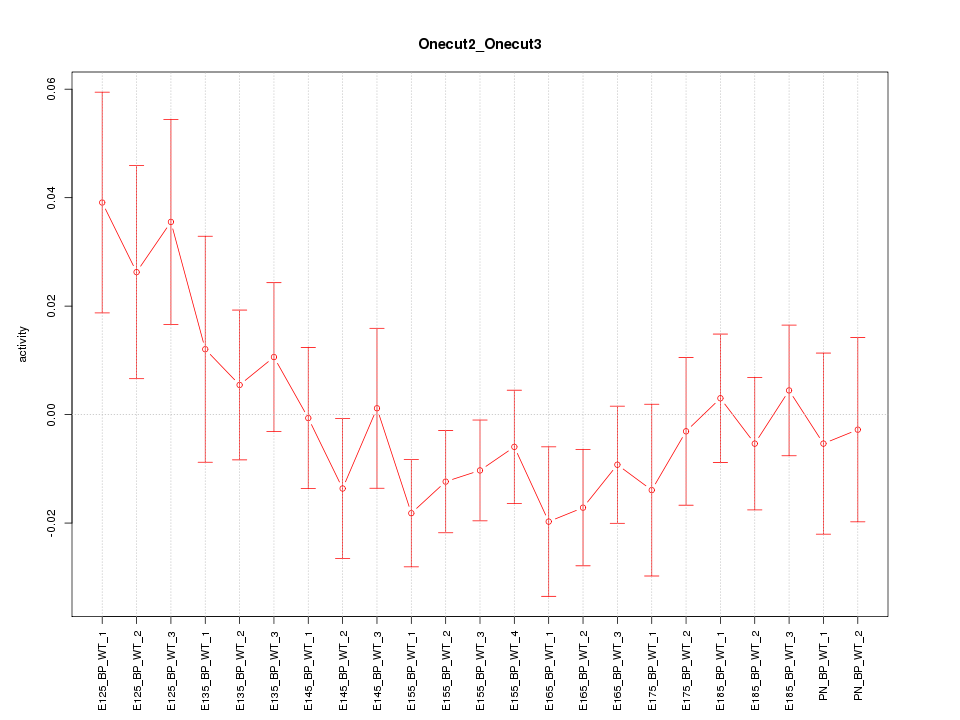
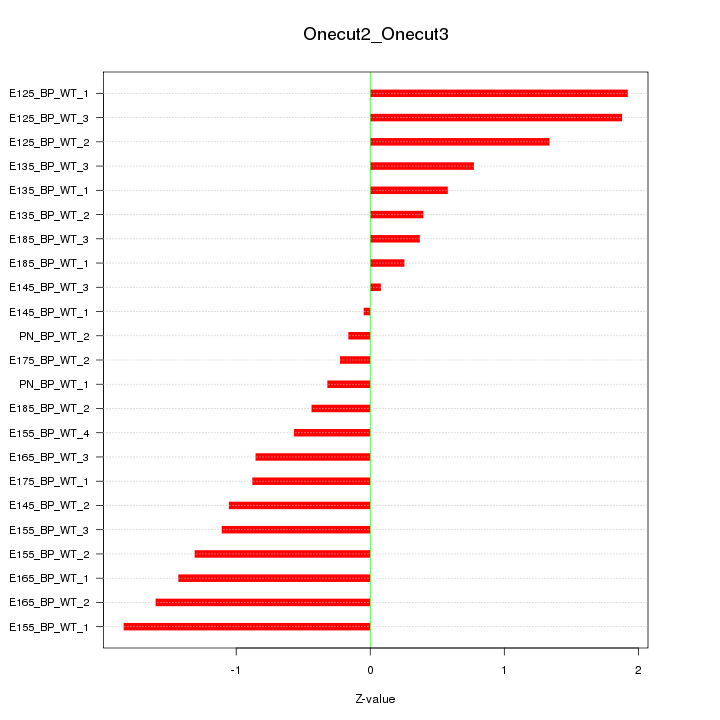
Motif ID: Onecut2_Onecut3
Z-value: 1.033
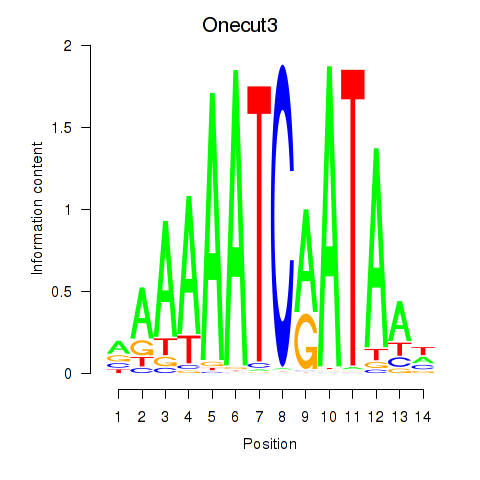
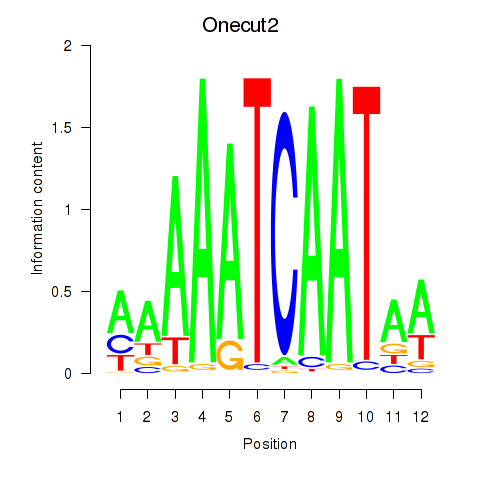
Transcription factors associated with Onecut2_Onecut3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Onecut2 | ENSMUSG00000045991.12 | Onecut2 |
| Onecut3 | ENSMUSG00000045518.8 | Onecut3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut3 | mm10_v2_chr10_+_80494835_80494873 | 0.62 | 1.7e-03 | Click! |
| Onecut2 | mm10_v2_chr18_+_64340225_64340364 | -0.40 | 6.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.7 | 3.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.6 | 10.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 2.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 9.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 3.9 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 1.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 3.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 2.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.9 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.0 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 0.6 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 0.6 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 1.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.3 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.1 | 2.9 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.8 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.3 | GO:1904008 | cellular response to salt(GO:1902075) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.1 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 0.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 1.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.9 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.0 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.2 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 3.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.4 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 3.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 1.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0030894 | replisome(GO:0030894) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.3 | 2.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.7 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 14.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 3.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 3.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 3.7 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.1 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |