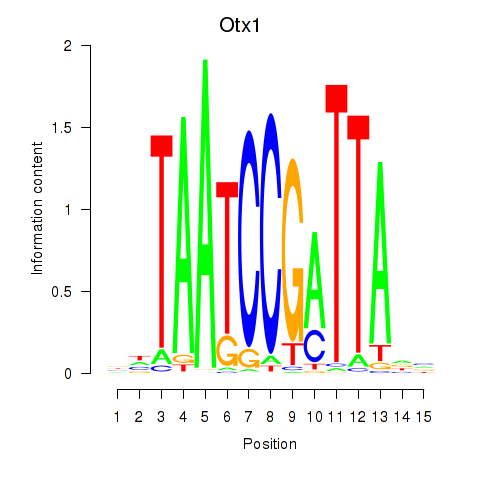
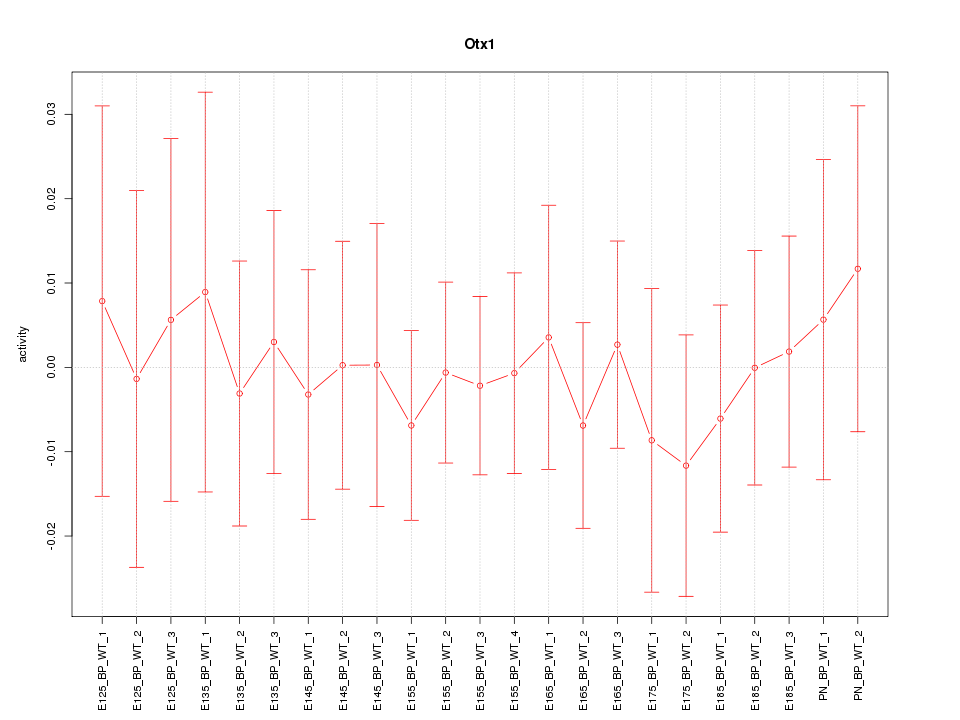
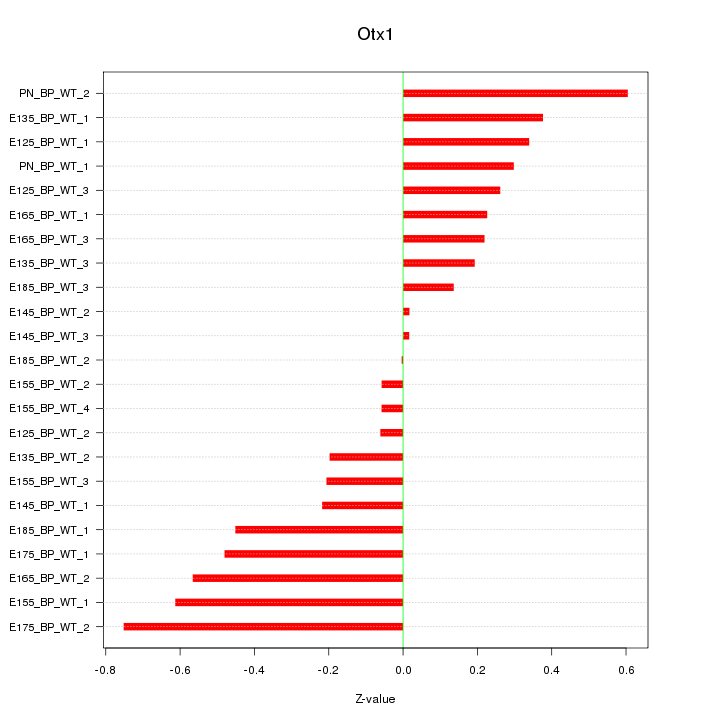
| 0.1 |
0.5 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.1 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) |
| 0.1 |
0.3 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 |
0.3 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.1 |
0.3 |
GO:0097503 |
sialylation(GO:0097503) |
| 0.0 |
0.1 |
GO:1900041 |
negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 |
0.3 |
GO:0071474 |
cellular hyperosmotic response(GO:0071474) |
| 0.0 |
0.2 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 |
0.1 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.2 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) membrane fission(GO:0090148) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 |
0.3 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.1 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 |
0.3 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.1 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.3 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.1 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.2 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.1 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.1 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |