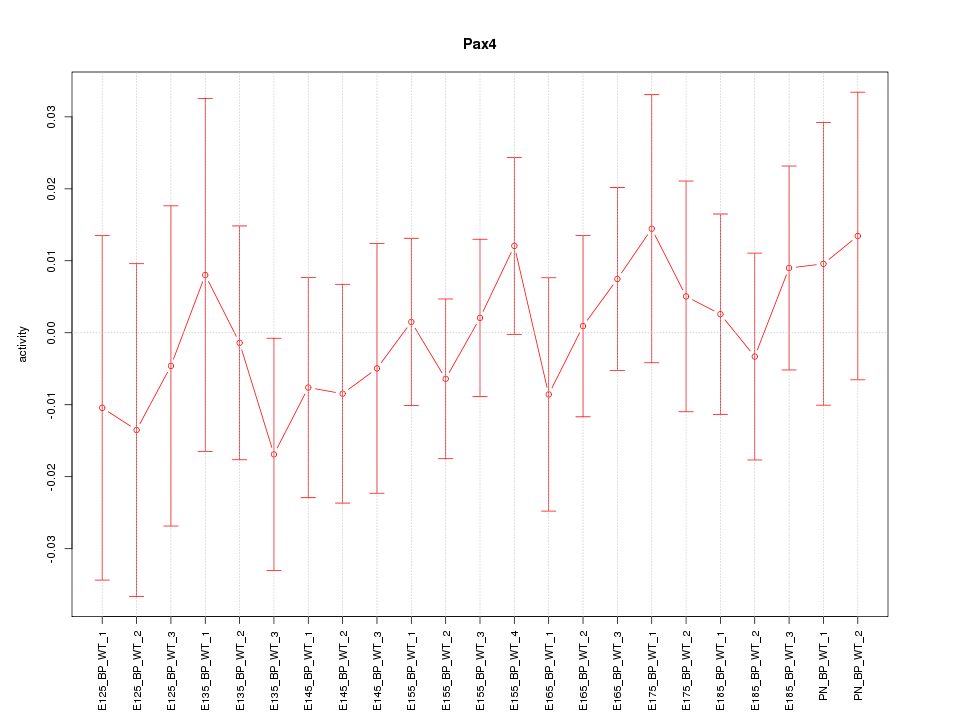
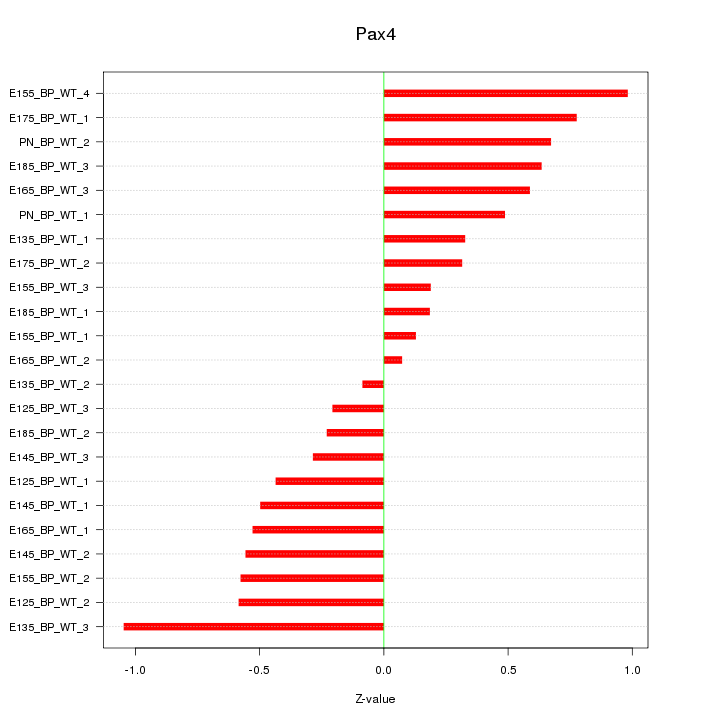
Motif ID: Pax4
Z-value: 0.523
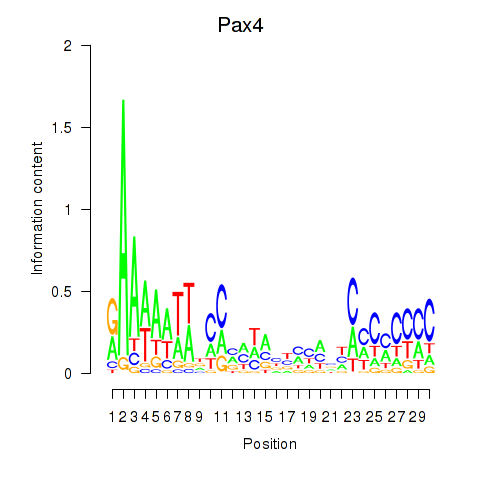
Transcription factors associated with Pax4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax4 | ENSMUSG00000029706.9 | Pax4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 0.9 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.5 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 1.0 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.6 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.0 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 1.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.6 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0016631 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.1 | 0.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 1.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |