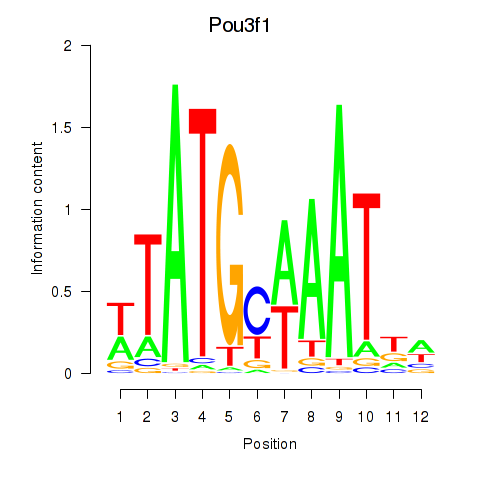
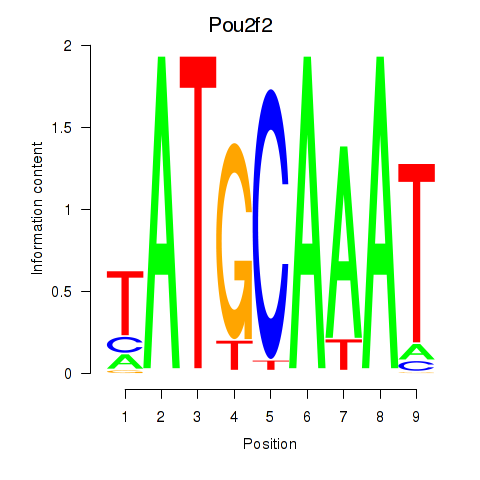
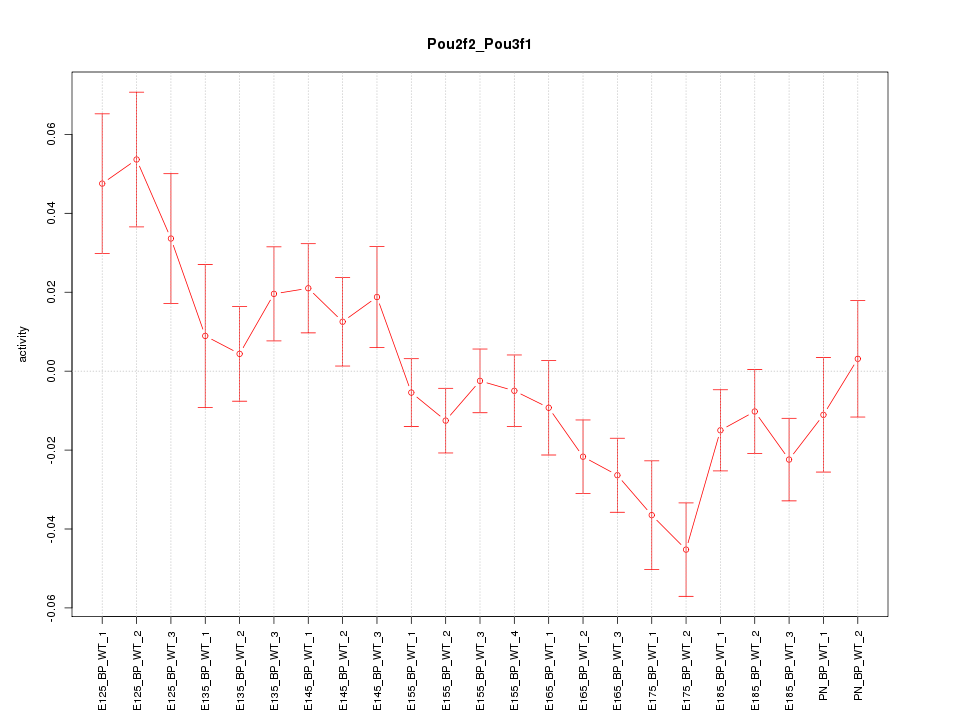
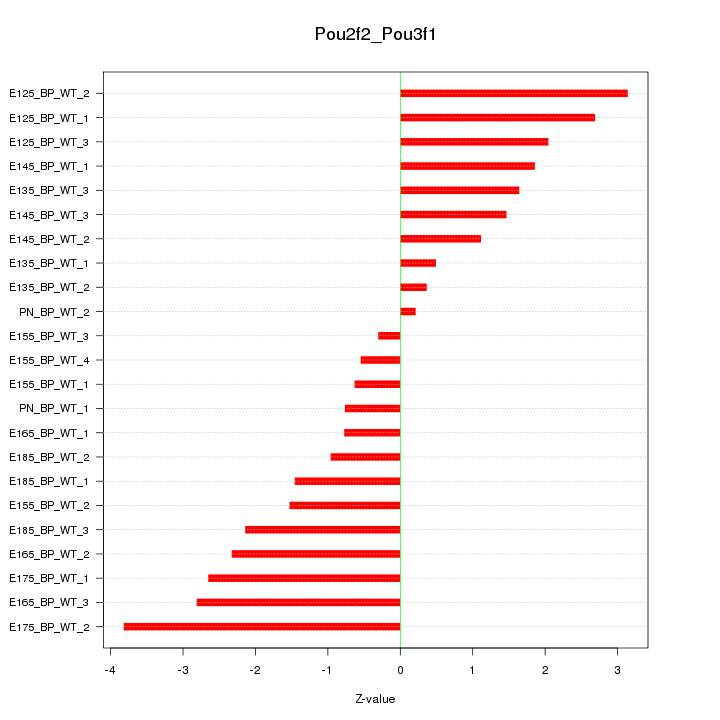
| 2.1 |
6.2 |
GO:0060853 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 2.1 |
6.2 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 1.9 |
7.6 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.7 |
8.6 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 1.5 |
22.6 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 1.3 |
9.0 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 1.2 |
3.5 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.1 |
13.6 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 1.1 |
14.5 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 1.1 |
2.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 1.0 |
5.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.0 |
2.9 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 1.0 |
30.9 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 1.0 |
3.8 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.9 |
7.2 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.7 |
2.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 |
4.1 |
GO:0097491 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.6 |
2.4 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.6 |
2.4 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.5 |
0.5 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.5 |
1.5 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 |
7.9 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.5 |
3.2 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.4 |
3.0 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 |
2.9 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.4 |
3.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 |
2.2 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 |
1.4 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.3 |
2.7 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 |
2.3 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 |
1.6 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.3 |
1.5 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 |
26.8 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.2 |
0.7 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 |
3.5 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.2 |
4.2 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.2 |
1.1 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 |
9.1 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.2 |
1.2 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
0.7 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.2 |
3.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 |
1.0 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 |
2.7 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.2 |
0.5 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 |
1.9 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.2 |
1.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 |
1.2 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
2.4 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
13.3 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.1 |
1.0 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 |
0.4 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.4 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
0.7 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.7 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
0.8 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 |
2.9 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
1.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.6 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.6 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 |
1.8 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
1.6 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 |
0.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
1.2 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
1.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.4 |
GO:0001696 |
gastric acid secretion(GO:0001696) oxalate transport(GO:0019532) |
| 0.1 |
0.2 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
2.6 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.1 |
0.9 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.5 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
1.3 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
1.8 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.3 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.2 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.9 |
GO:0030262 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 |
2.0 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
2.2 |
GO:0032330 |
regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 |
8.9 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
1.4 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 |
0.0 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.0 |
1.3 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
1.1 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.0 |
0.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.5 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.3 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 |
0.4 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
2.3 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
2.1 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
2.3 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.4 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.6 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
1.1 |
GO:1903169 |
regulation of calcium ion transmembrane transport(GO:1903169) |