Motif ID: Pou3f3
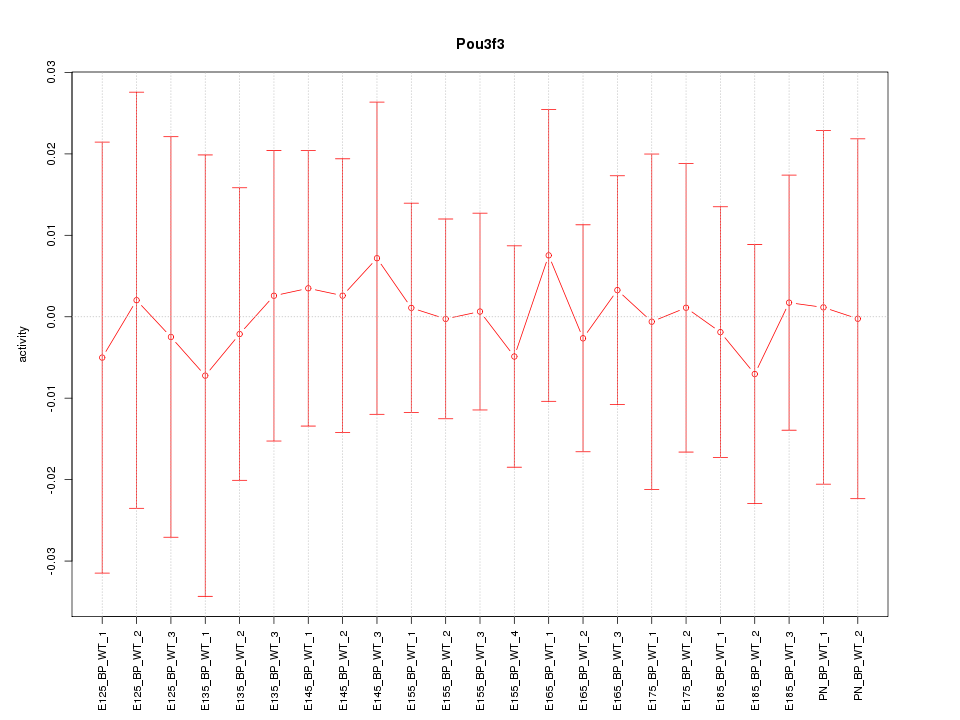
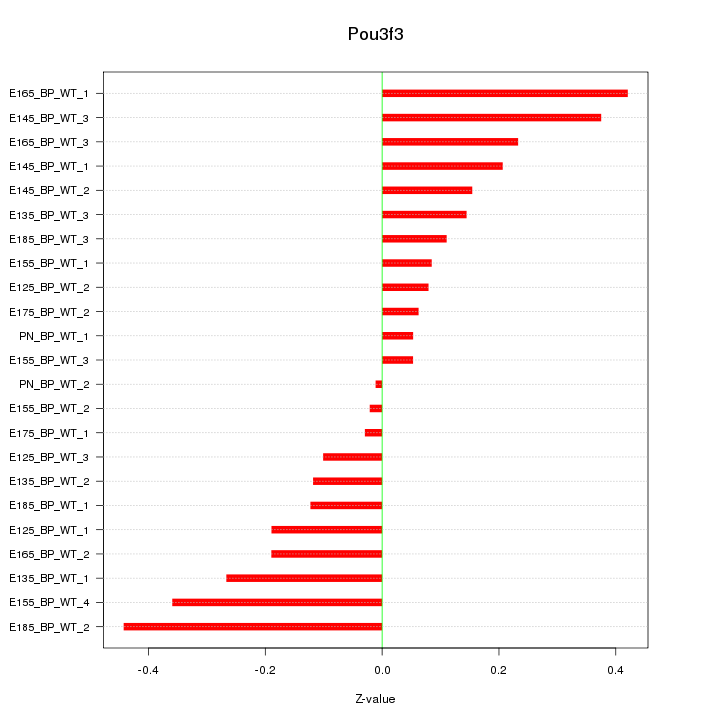
Z-value: 0.209
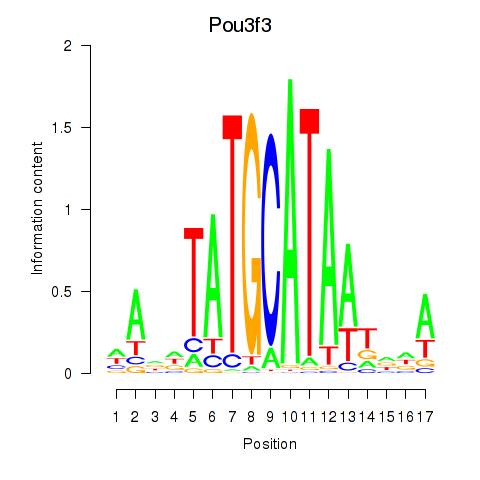
Transcription factors associated with Pou3f3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou3f3 | ENSMUSG00000045515.2 | Pou3f3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f3 | mm10_v2_chr1_+_42697146_42697146 | -0.04 | 8.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904956 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |